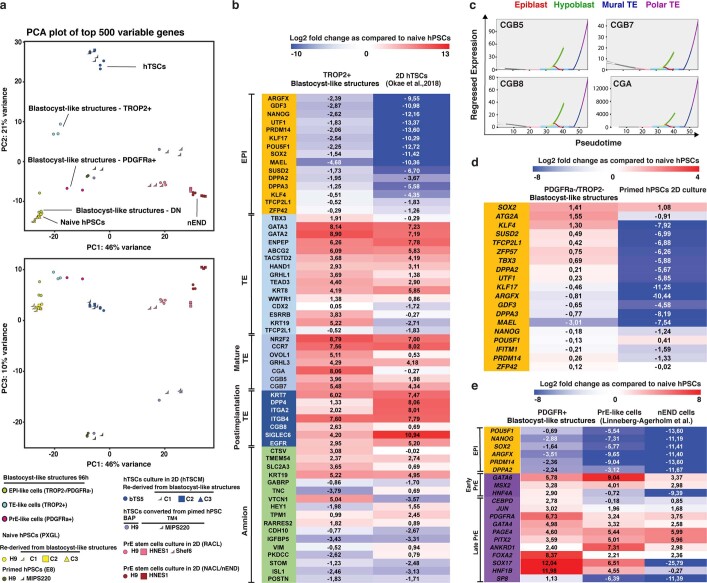
Extended Data Fig. 4. Cells in human blastocyst-like structures are transcriptionally similar to pre-implantation lineages.
a. Principal component analysis (PCA) plot with PC1 vs PC2 (top) or PC1 vs PC3 (bottom) computed with top 500 variable gene in the bulk transcriptome of individual lineages of blastocyst-like structures (EPI, TE and PrE); stem cell lines: naive and primed hPSCs; hTSCs: blastocyst derived hTSCs (bTS5)20, primed hPSC derived hTSCs (BAP14 and TM4 protocols45; PrE like stem cell lines (RACL or nEND cells22); naive PSC and TSCs rederived from blastocyst-like structures (see methods). b. Heatmap of key blastocyst and post-implantation lineage markers differentially expressed between TE analogs (TROP2+) of the blastocyst-like structures and hTSCs in their bulk transcriptome. c. Pseudotime analysis of human mature TE markers CGB5, CGB7, CGB8 and CGA. Gene expression analysis was performed by using the public data analysis tool (https://bird2cluster.univ-nantes.fr/demo/PseudoTimeUI/). d. Heatmap of key pluripotency related genes differentially expressed between EPI analogs (PDGFR-/ TROP2-) in the blastocyst-like structures and primed hPSCs e. Heatmap of key pluripotency related genes or PrE markers differentially expressed between PrE analogs (PDGFRα+) in the blastocyst-like structures, naive PSC derived PrE-like cells and nEND cells.