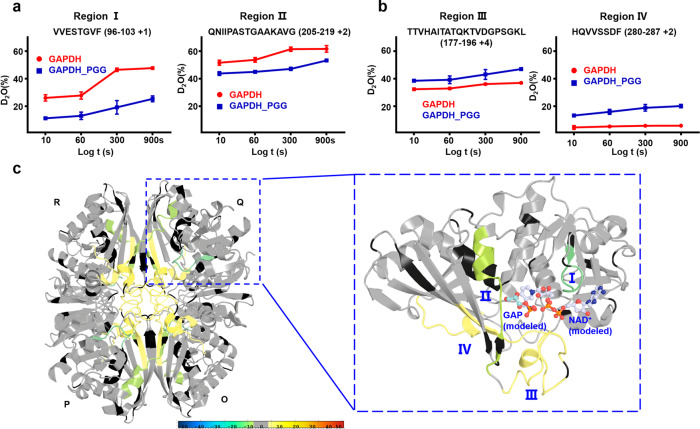
Fig. 3. HDX-MS revealed the binding region and conformation change area of GAPDH upon PGG binding.
a The deuterium uptake plot for region I and region II are shown. Deuterium uptake plot of the peptide VVESTGVF (AA 96–103, +1 charge state) (region I) and QNIIPASTGAAKAVG (AA 205–219, +2 charge state) (region II) in GAPDH in the presence and absence of PGG are shown. The data are plotted as percent deuterium uptake versus time on a logarithmic scale. Red and blue plots represent the GAPDH and the PGG bound state, respectively. b The deuterium uptake plot for region III and region IV are shown. Deuterium uptake plot of the peptide TTVHAITATQKTVDGPSGKL (AA 177–196 +4) (region III) and HQVVSSDF (AA 280–287 +2) (region IV) for GAPDH in the presence and absence of PGG are shown. The data were plotted as percent deuterium uptake versus time on a logarithmic scale. Red and blue plots represent the GAPDH and the PGG bound state, respectively. c Differential HDX consolidation view (related to Supplementary Fig. S3A and S3B) was mapped to + the GAPDH crystal structure (PDB ID: 1U8F). The GAPDH monomer is shown on the right, the enlarged view was also modeled with GAP and NAD+, NAD+ was modeled by 1U8F, the modeled GAP was based on the PDB ID: 1NQO of Bacillus stearothermophilus GAPDH. HDX Workbench colors each peptide according to the smooth color gradient HDX perturbation key (D%), as shown in each figure. Differences in %D between −5% and 5% are considered non-significant and are colored gray according to the HDX perturbation key shown below. In addition to the −5% to 5% test, unpaired t tests were calculated to determine significant differences (P < 0.05) between samples at each time point. A negative value represents decreased deuterium incorporation or stabilization, while a positive value represents increased deuterium incorporation or destabilization in the corresponding region of the receptor when a binding event takes place. Peptides exhibiting statistically insignificant or undetectable changes are colored gray. The blank region represents undetected peptide for the corresponding experiment.