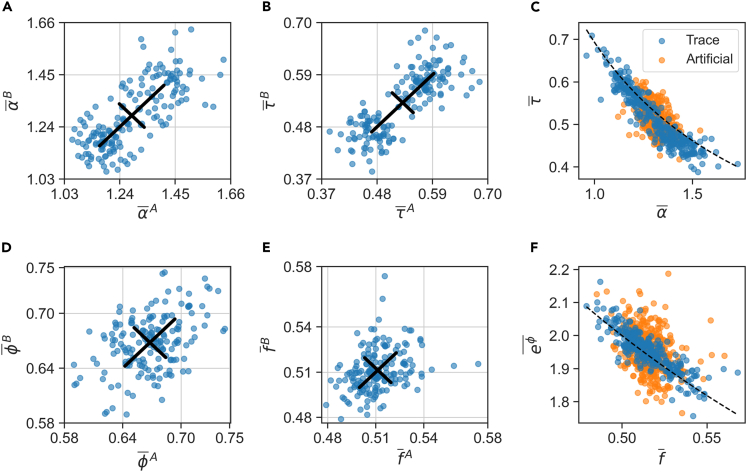
Figure 5.
Covariation of sloppy homeostatic set points
(A and B) Sloppy variables α, τ display a range of homeostatic set points (time averages). Scatter plots for two neighboring lineages in the same microfluidic trap: (A) Exponential growth-rate α, Pearson ; (B) generation time τ, . The set points are primarily sensitive to the environment (diagonal spread), and less so to lineage history (off-diagonal spread). Black lines are standard deviations in the two directions.
(C) Within each lineage, set points strongly covary (blue circles; ) and lie close to the line (dashed black). Orange circles show averages of per-cycle variables over arbitrary groups of the same sizes as lineages (artificial lineages; ).
(D) Scatter plot of fold-growth set points ϕ for neighbor cells, Pearson .
(E) Scatter plot of division fraction set points for neighbor cells, Pearson .
(F) Same presentation as in C, for the variables and . The dashed black line is ; for temporal averages, for artificial averages. For a full matrix of set point scatter plots for pairs of variables between neighbor lineages, see Figure S7