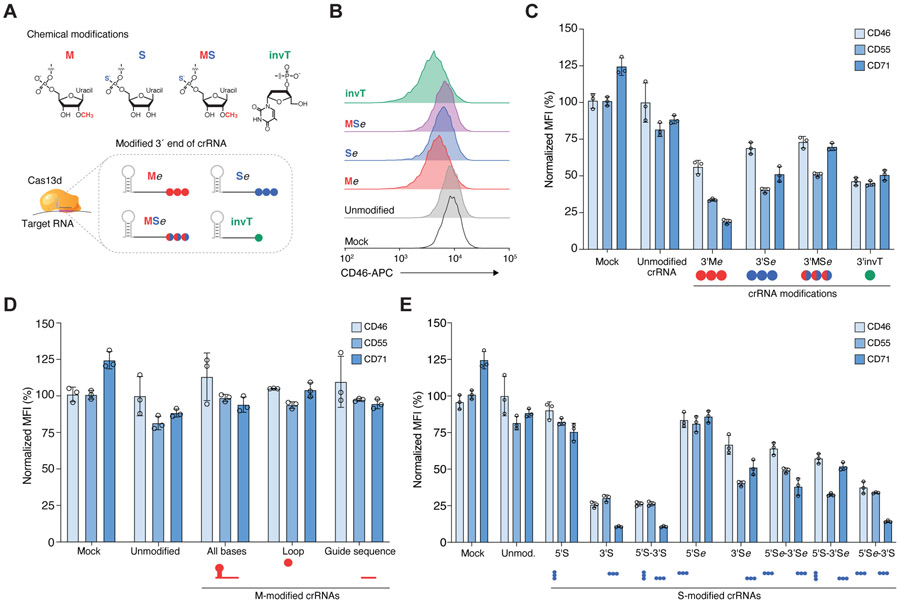
Figure 1. Chemically-modified CRISPR RNAs (crRNAs) improve Cas13 knockdown efficacy in human cells.
(A) Overview of chemical modifications incorporated during synthesis of crRNAs: M, 3′-O-methyl base; S, phosphorothioate bond; MS, 3′-O-methyl base and phosphorothioate bond; invT, inverted thymidine. All crRNA sequences and modifications are listed in Table S1. (B) Representative flow cytometry results for CD46 knockdown in HEK293FT-TetO-RfxCas13d-NLS cells nucleofected with synthetic crRNAs targeting CD46 — specifically, three modified uridines (Me, Se, MSe modifications with e denoting an extended crRNA with extra uridines) or an inverted thymidine directly following the last base of the guide sequence (invT modification). (C, D, E) Percent of CD46, CD55 and CD71 protein expression 72 hours post-nucleofection with the indicated crRNAs in HEK293FT-TetO-RfxCas13d-NLS cells. Cas13 expression was induced with doxycycline 24 hours prior to crRNA nucleofection. Target protein expression was measured by flow cytometry. RNA modification and placement were as follows: (C) synthetic crRNAs with chemical modifications located in the 3’ crRNA end — specifically, three modified uridines (M, S, MS modifications) or directly following the last base of the guide sequence (invT modification); (D) synthetic crRNAs with 3′-O-methyl (M) modification in all nucleotides in the crRNA (direct repeat and guide), in 3 nt in the loop region of the direct repeat, or in the 23 nt guide sequence; and (E) synthetic crRNAs chemically modified with a phosphorothioate bond placed in the crRNA sequence at the 5’ and/or 3’ end (S), or in three uridines added at the 5’ and/pr 3’ end (Se). Relative protein expression was calculated by comparing the normalized median fluorescent intensity (MFI) to cells nucleofected with non-targeting (NT) crRNAs. Bars represent mean values ± s.d., n = 3 biological replicate nucleofections.