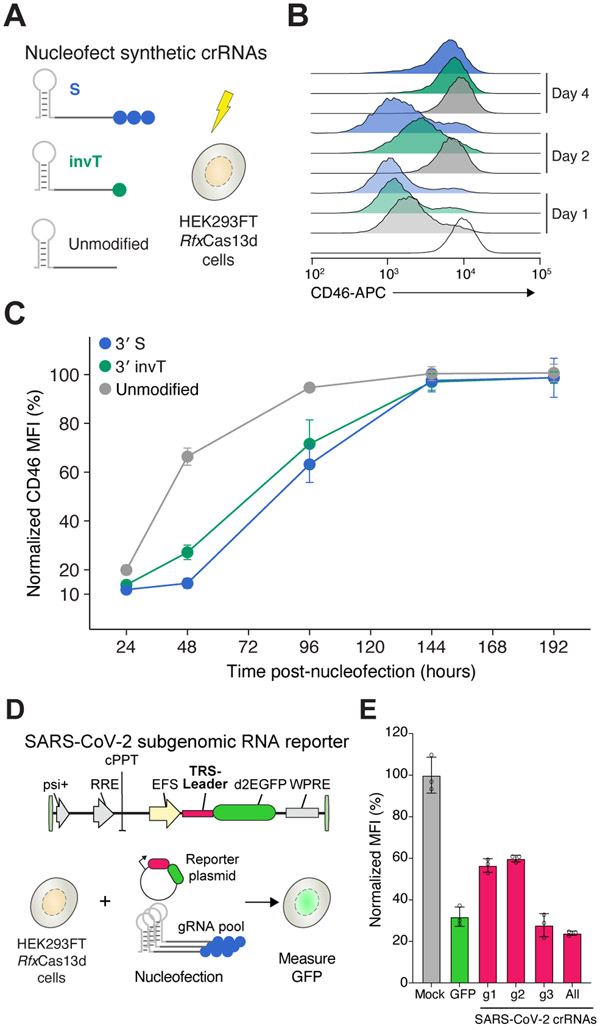
Figure 2. Transcript knockdown with chemically-modified crRNAs is sustained over multiple days.
(A) Experimental design to measure the temporal dynamics of transient CD46 knockdown by nucleofecting the synthetic crRNAs in the HEK293FT-TetO-RfxCas13d-NLS cells. Synthetic crRNAs were unmodified, chemically modified with a phosphorothioate bond (S) on three uridines at the 3’ end, or chemically modified with an inverted thymidine at the 3’ end (invT). (B) Representative CD46 histograms at 1, 2 and 4 days after synthetic crRNA nucleofection. (C) Relative CD46 protein expression upon nucleofection with the synthetic crRNAs in panel A, normalized to cells nucleofected with non-targeting NT crRNAs. Points represent mean values ± s.d. , n = 3 biological replicate nucleofections. (D) Experimental approach to measure SARS-CoV-2 RNA knockdown using a subgenomic RNA fluorescent reporter (destabilized EGFP) with synthetic crRNAs chemically modified with a phosphorothioate bond (S) on the three terminal nucleotides of the guide sequence. (e) EGFP expression in HEK293FT-TetO-RfxCas13d-NLS cells. Nucleofection conditions: GFP, pool of three S-modified crRNAs targeting EGFP; g1, g2 and g3, individual S-modified crRNAs targeting the SARS-CoV-2 TRS-Leader sequence; All, pool of g1, g2 and g3 S-modified crRNAs. Percent GFP expression is determined relative to cells nucleofected with NT crRNAs. Bars represent mean values ± s.d. , n = 3 biological replicate nucleofections.