FIGURE 1.
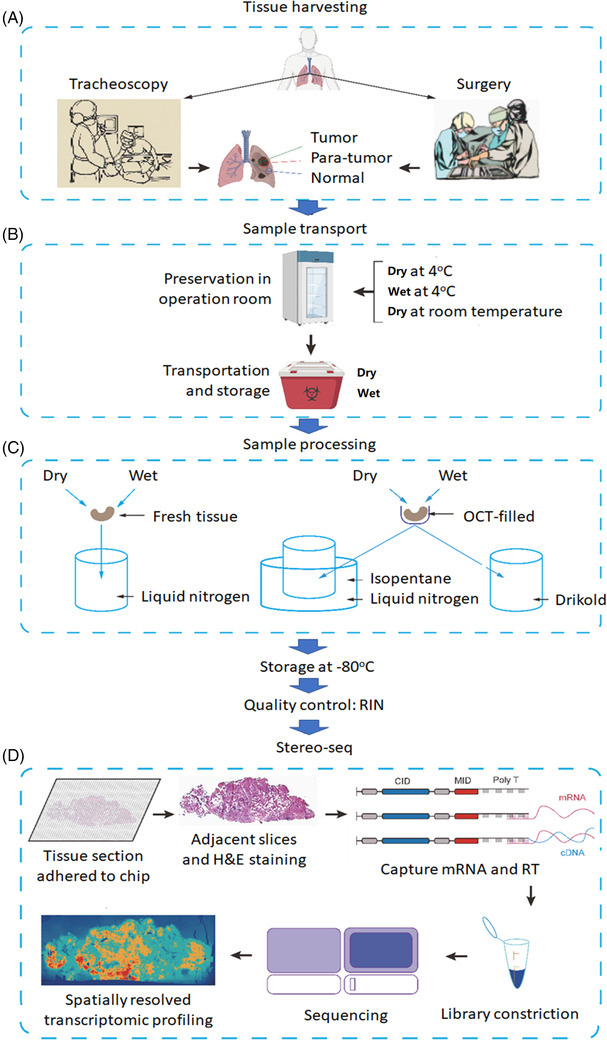
Processes and workflow of clinical spatial transcriptome from sample harvest to transcriptomic imprints. (A) Tissue harvesting: human samples of lung tumour, para‐tumour (>2 cm from cancer edge) and normal tissues (>3 cm from para‐cancer tissue) were harvested from either tracheoscopy or surgery. (B) Sample transport: the harvested tissues were temporarily maintained with physiological saline solution (‘wet’) or without solution (‘dry’) in the operating room at 4℃ or room temperature, then samples were transported to the laboratory from on ice. (C) Sample processing: tissues at wet or dry conditions were frozen directly in liquid nitrogen or were immersed with the solution of optimal cutting temperature compound (OCT) in isopentane cooled with liquid nitrogen or drikold. The processed samples were temporarily stored in the refrigerator at –80℃, and then RNA integrity number (RIN) value was detected. (D) Stereo‐seq: Spatio‐Temporal Enhanced Resolution Omics‐sequencing (Stereo‐Seq) is an emerging next generation of spatially resolved transcriptomics technology using the high resolution of regularly and septically spaced DNA nanobeads (DNB) to enable in situ RNA capture, amplification and sequencing
