Figure 1. Single-nucleotide detection of dihydrouridine (D) by rhodamine labeling.
(A) Comparative Rho (rhodamine) detection at 520 nm on WT (wild-type) and Δ4 (Δdus1–2-3–4) total RNAs in R+ and mock-treated (R−) samples. Intensity of a Rho drop is scaled at 100%, a drop of water is considered as the background, and methylene blue staining serves as loading control. n = 3 biological replicates; error bars represent SEM.
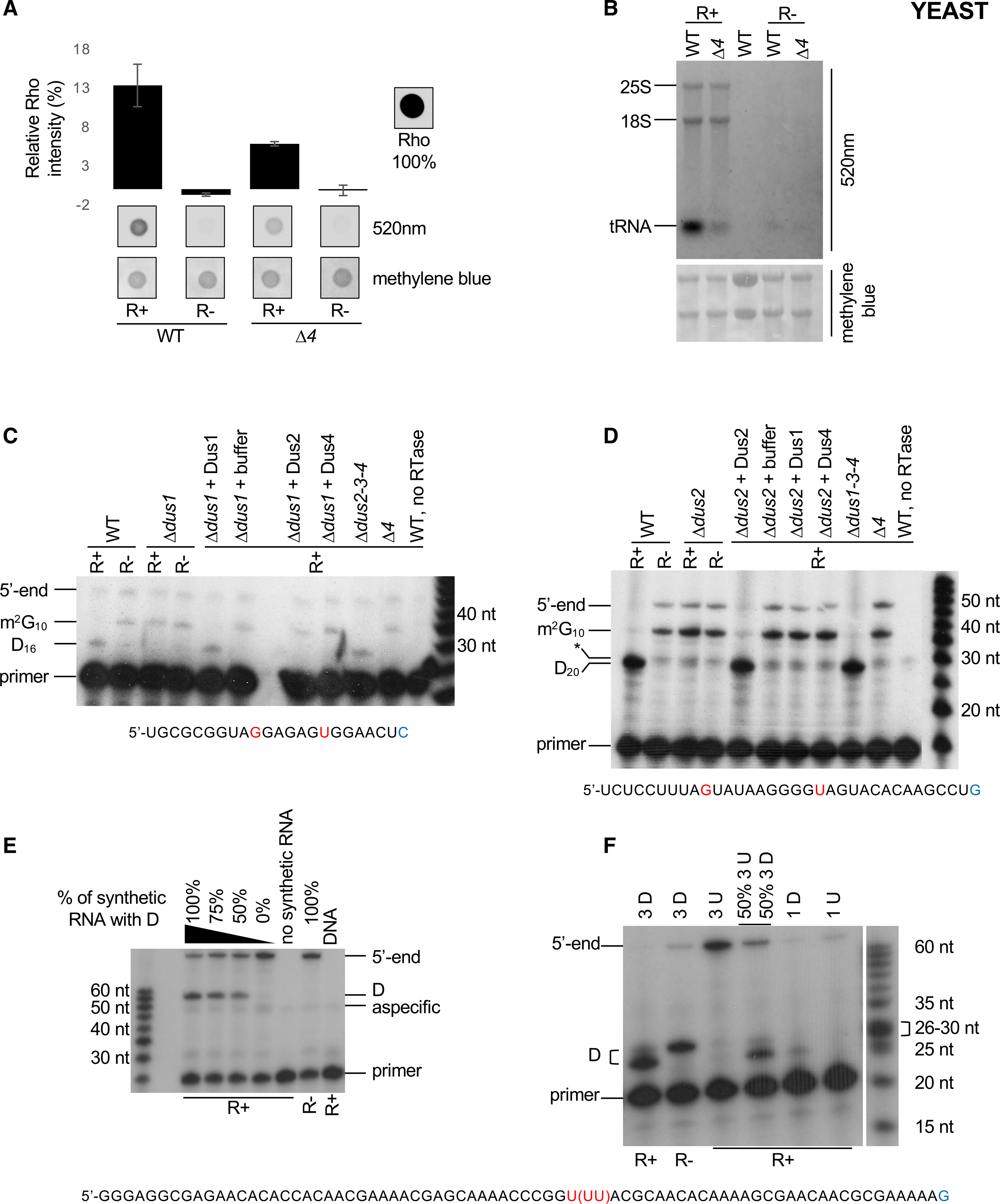
(B) Fluorescent northern blotting of WT and Δ4 total RNAs in R+ and mock-treated samples. Untreated total RNA from WT strain is shown in lane 3. Representative result from a biological triplicate; methylene blue staining serves as loading control.
(C) Primer extension assays specific to tRNAiMet (Dus1-dependent D16) were performed on total RNA from WT, single dus (Δdus1), triple dus (Δdus2–3-4), or quadruple dus (Δ4) mutant strains, R+ or R− treated as indicated. Before the assay, in vitro dihydrouridylation reactions were performed on total RNA with GST-fusion S. cerevisiae proteins (+ Dus1, 2, or 4) or buffer. G10 and U16 are highlighted in red, and the last nucleotide of the primer is in blue. m2G10 indicates a labeling-independent RT stop.
(D) Primer extension assays specific to tRNAAspGUC (Dus2-dependent D20) performed on total RNA from WT, single dus (Δdus2), triple dus (Δdus1–3-4), or quadruple dus (Δ4) mutant strains, R+ or R− treated as indicated. Before the assay, in vitro dihydrouridination reactions were performed on total RNA with GST-fusion S. cerevisiae proteins (+ Dus1, 2, or 4) or buffer. G10 and U20 are highlighted in red, and the last nucleotide bound by the 3′ end of the primer id in blue. m2G10 indicates a labeling-independent RT stop, and the asterisk indicates a nonspecific band.
(E) Primer extension assay was performed on an in vitro-transcribed RNA containing 1 U/D mixed with S. pombe total RNA. A decreasing ratio of the dihydrouridinated RNA was added before R+ or R− labeling reactions. DNA is the PCR product used for in vitro transcription. The unique U43 is highlighted in red, and the last nucleotide bound by the 3′ end of the primer is in blue.
(F) Primer extension assay was performed on an in vitro-transcribed RNA mixed with S. pombe total RNA before R+ or R− labeling reactions. 3 D indicates an in vitro RNA containing three consecutive D residues at position 25, 26, and 27, respectively. 3 U indicates an in vitro-transcribed RNA where the corresponding positions are uridine. 1 D and 1 U are the same synthetic RNAs used in (E). The triplet U43−45 is highlighted in red, and the last nucleotide bound by the 3′ end of the primer is in blue.
