Figure 7. D is detected on the tubulin mRNA in human cells.
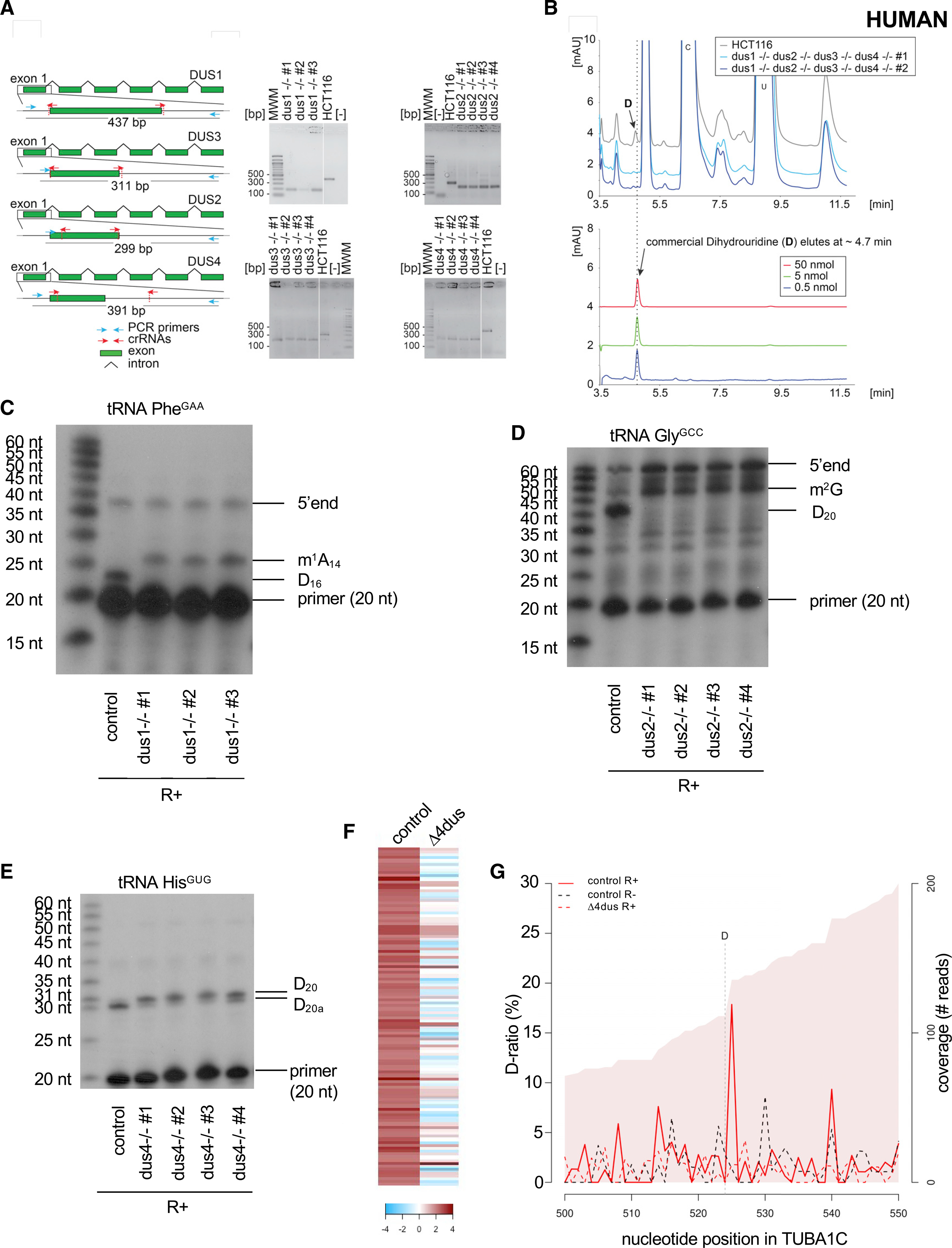
(A) Left: structure of the genomic loci of the four human DUS genes. For all four DUSes, exon 1 containing the catalytic domain was targeted for precise deletion by CRISPR-Cas9 in HCT116 diploid cells. The guide RNAs (crRNAs) used are depicted as red arrows. The primers used for the diagnostic PCR are highlighted in cyan, and the size of the PCR-amplified product is indicated. Right: diagnostic of the cell lines by PCR. Genomic DNA extracted from the indicated purified clones and isogenic reference control (HCT116) was subjected to a PCR amplification with indicated primers (blue arrows). MWM, molecular weight marker; [−], control without DNA. Note the site of deletion was confirmed by DNA sequencing for all cell lines. In all cell lines, both alleles were deleted.
(B) Total RNA extracted from human cells lacking all four DUS genes and from isogenic reference control cells (HCT116) was analyzed by quantitative HPLC for the presence of D. Lower panel: synthetic D was used for calibration (the indicated amounts were used).
(C) Primer extension assays specific to human tRNAPhe(GAA) (containing a Dus1-dependent D16) were performed on total RNA from control cells (HCT116) and three clones of the ΔDUS1. m1A14 and the 5′ end are indicated.
(D) Primer extension assays specific to human tRNAGly(GCC) (containing a Dus2-dependent D20) were performed on total RNA from control cells (HCT116) and four clones of the ΔDUS2. m2G and the 5′ end are indicated.
(E) Primer extension assays specific to human tRNAHis(GUG) (containing a Dus4-dependent D20a and a Dus2-dependent D20) were performed on total RNA from control cells (HCT116) and four clones of the ΔDUS4.
(F) Heatmap highlighting the 112 D-sites detected within human mRNAs sorted by comparison of their respective D-ratios in test (R+) and control (R−) conditions. Dark red highlights more RT terminations in the test condition (D-ratio R+ > D-ratio R−). The columns depict the cell lines used.
(G) Evolution of the D-ratio (%) and the coverage (reads) in the indicated conditions along the TUBA1C (tubulin alpha 1C) mRNA. The R+ reaction generates a high (about 17%) D-ratio coupled to a decrease of coverage, which supports the presence of a D position one nucleotide upstream.
