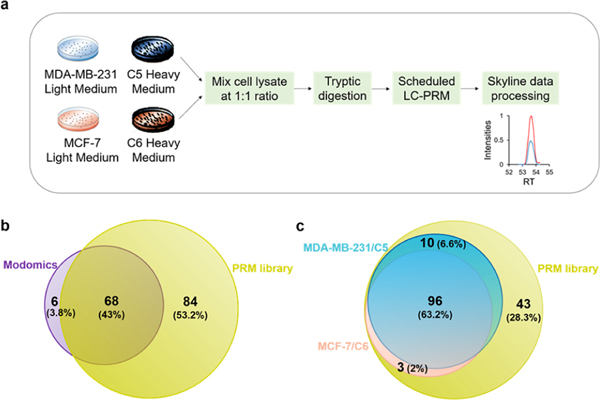
Figure 1.
LC–PRM method for uncovering alterations in expression of epitranscriptomic RWE proteins associated with the development of radioresistance. (a) A SILAC-based LC–PRM workflow. The parental cells (i.e., MDA-MB-231 and MCF-7) and their radioresistant counterparts (i.e., C5 and C6) were labeled in light- or heavy- amino acid-containing media for over six cell doubling times. In the forward SILAC labeling experiments, light-isotope-labeled C5 and C6 cell lysates were mixed at a 1:1 ratio (by mass) with heavy-isotope-labeled MDA-MB-231 and MCF-7 cell lysates, respectively. In the reverse SILAC labeling experiments, light-isotope-labeled MDA-MB-231 and MCF-7 cell lysates were mixed at a 1:1 ratio (by mass) with heavy-isotope-labeled C5 and C6 cell lysates, respectively. The mixed cell lysate was tryptic digested and subjected to LC–PRM analysis. Data were processed using Skyline. (b,c) Venn diagrams showing the number and percentage of human epitranscriptomic RWE proteins deposited in the Modomics database (purple) compared with those included in the PRM library of this study (yellow) (b) and illustrating the number and percentage of quantified epitranscriptomic RWE proteins in MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells from LC–PRM analyses, compared with those deposited in the PRM library (c). Blue and pink circles in parts b and c designate the numbers of quantified epitranscriptomic RWE proteins in MDA-MB-231/C5 and MCF-7/C6 pairs of breast cancer cells, respectively.