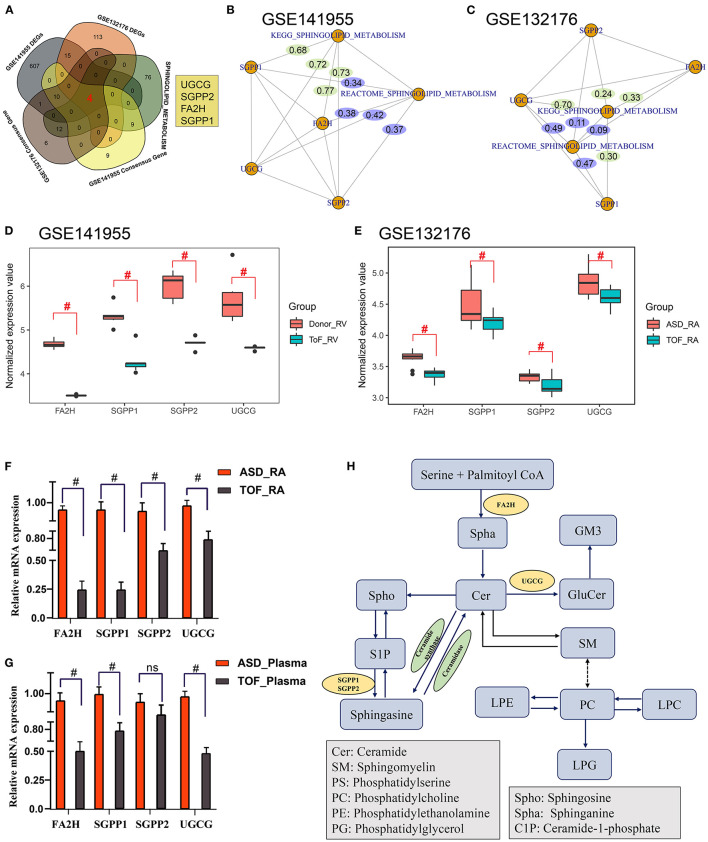
Figure 7.
Identification of core regulatory genes related to sphingomyelin metabolism in TOF. (A) Common genes among the GES141955 differentially expressed genes (DEGs), GSE132176 DEGs, GES141955 consensus genes, GES132176 consensus genes, and sphingomyelin metabolism-related genes. (B) Gene-pathway interaction network by partial correlation analysis based on standardized gene expression values and sphingomyelin metabolic pathway scores in the GSE141955 dataset. (C) Gene-pathway interaction network by partial correlation analysis based on standardized gene expression values and sphingomyelin metabolic pathway scores in the GSE132176 dataset. (D,E) Expression of core genes in GSE132176 and GSE141955. (F,G) QT-PCR validation of the expression of the four core genes. (H) Mechanism plot of the study. #p < 0.05.