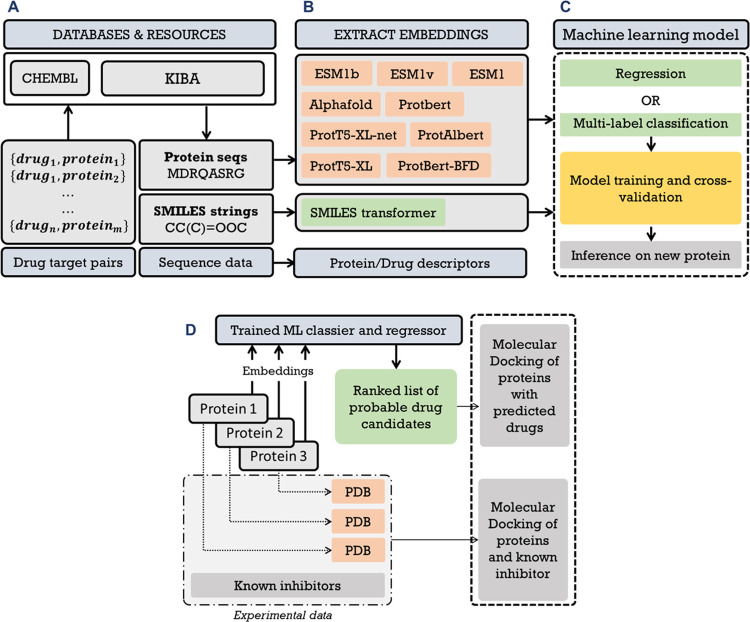
Figure 1.
General overview of the overall methodology. (A) Primary DTI data were collected, processed, and screened. (B) Embeddings were generated for protein and drug sequences using multiple language model-based methods. (C) Extracted embeddings were used to train a fully connected feedforward deep neural network for classification and regression task. (D) Molecular docking and dynamics workflow for validating the activity of predicted drug–target interactions.