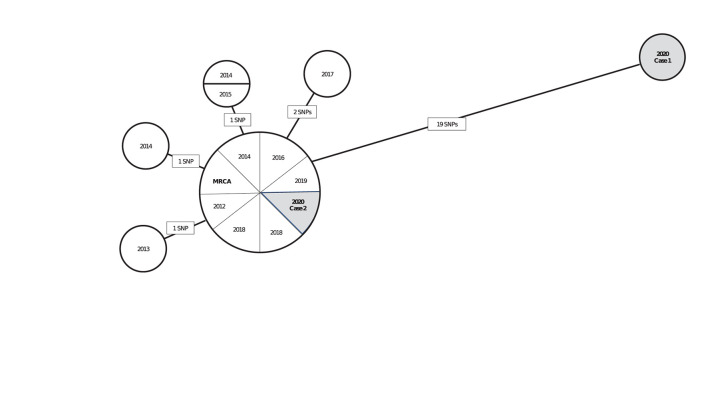
Figure 1.
A hypothetical neighbor-joining tree (phylogenetic analysis) representing the genetic distances in single nucleotide polymorphisms (SNPs) among 15 isolates of Mycobacterium tuberculosis (Mtb) from culture-confirmed tuberculosis cases reported during 2010–2020a. aIsolates are displayed as circles called nodes; isolates with the same genome sequence are displayed together in one node. Lines between nodes are labeled with the number of SNPs (mutations at a single position in the DNA sequence), and these lines are proportional in length to the number of SNPs. The most recent common ancestor (MRCA) is a hypothetical genome (not an actual isolate) from which all isolates in the phylogenetic analysis are descended. The MRCA serves as a reference point for examining the direction of genetic change. In this hypothetical scenario, a TB control program is using these phylogenetic analysis results to help determine if isolates from two patients reported during 2020 (shaded in gray) are likely attributable to recent transmission. If so, those cases are a priority for further investigation. Interpretation of these results, depending on if the Mtb mutation rate is assumed to be similar during latency or to be slower, is as follows: Case 1 is unlikely to be involved in recent transmission under either assumption because the patient's isolate is genetically distant to that of the patient in Case 2 and all other cases in the analysis (≥19 SNPs). This interpretation might change as new cases are reported and added to the phylogenetic analysis. Case 2 is more challenging to interpret: under an assumption that Mtb mutates at a similar rate during latent infection and disease, Case 2 is likely to be involved in recent transmission because the patient's isolate is genetically close to those of other cases in the analysis (0–2 SNPs). If Case 2 was attributable to reactivation after Mtb infection during the remote past, more SNPs can be expected. Under an assumption that Mtb mutates at a slower rate during latency than disease, Case 2 might be involved in recent transmission or attributable to reactivation after Mtb infection during the remote past because relatively few SNPs are expected to accumulate during latency. Other clinical and epidemiologic data are needed for making a determination.