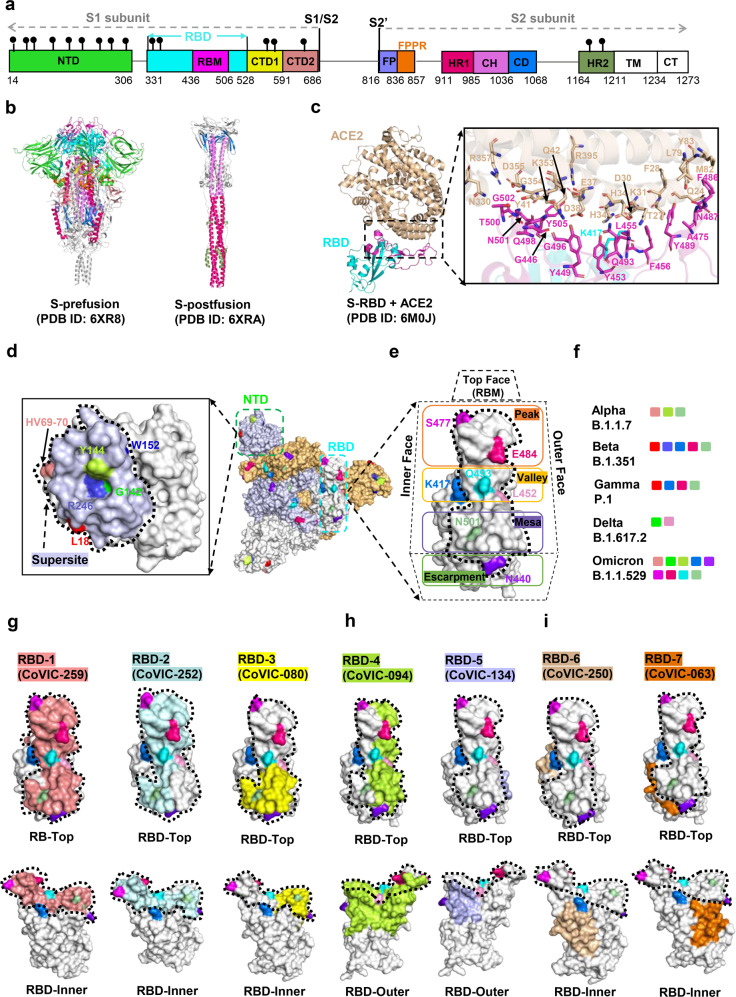
Fig. 2.
Structures of SARS-CoV-2 Spike protein and antibody recognition. a The full-length SARS-CoV-2 S protein. NTD N terminal domain, RBD receptor-binding domain, RBM receptor-binding motif, CTD1 C-terminal domain 1, CTD2 C-terminal domain 2, FP fusion peptide, FPPR fusion-peptide proximal region, HR1 heptad repeat 1, CH central helix region, CD connector domain, HR2 heptad repeat 2, TM transmembrane segment, CT cytoplasmic tail. b Structures of S trimer in prefusion and postfusion states. Each domain is marked with a color corresponding to a. c Structure of the RBD (cyan) in complex with ACE2 (pink). Residues involved in interactions between the RBD and ACE2 are shown as sticks. d Footprints for NTD-targeted antibodies, with the NTD “supersite” outlined with a dashed line. The residue positions of important mutations and deletions are indicated in the NTD. Table 1 lists mutations represented in each variant. e Location of important emerging mutations on the RBD. The RBM can be topologically divided into three subsections: the “peak” that includes residues S477 and E484; the “valley” that includes K417, Q493, and L452; and the “mesa” includes N501. f Mutations and deleted residues affecting antibodies activity involved in significant mutants. g–i footprint of a representative antibody from the Coronavirus Immunotherapeutic Consortium (CoVIC) mapped onto an RBD monomer. The ACE2 binding site is outlined with a dotted line. The website of CoVIC is at https://covic.lji.org/