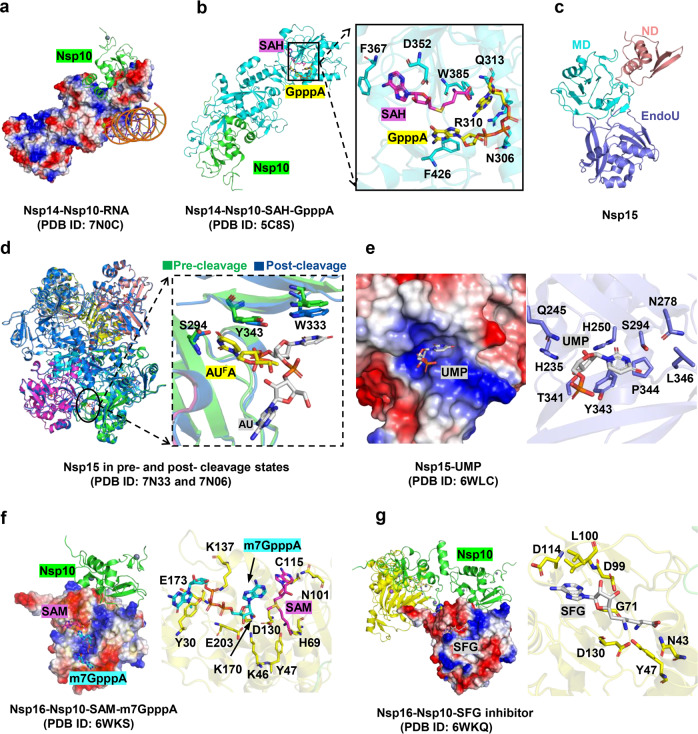
Fig. 7.
Structures of Nsp14 /Nsp15/Nsp16 and their inhibitors. a Cryo EM structure of the SARS-CoV-2 Nsp10-Nsp-14 RNA complex. Nsp14 is illustrated with electrostatic surface. Nsp10 is illustrated with cartoon in green. b The structure of the Nsp14-Nsp10 in complex with functional ligands S-adenosyl-L-homocysteine (SAH) and GpppA shown in sticks. c The overall structural of Nsp15. d Conformational changes between Nsp15 in pre- and post- cleavage states. e Structures of Nsp15 in complex with uridine-5′-monophosphate (UMP). f The structure of SARS-CoV-2 Nsp16-Nsp10 in complex with RNA cap analogue (m7GpppA) and S-adenosyl methionine (SAM). The m7GpppA and SAM are shown in sticks. Nsp16 is indicated as electrostatic surface. g The structure of Nsp16-Nsp10 heterodimer in complex with sinefungin (SFG). SFG is shown in sticks. Nsp16 is illustrated with electrostatic surface and cartoon in yellow