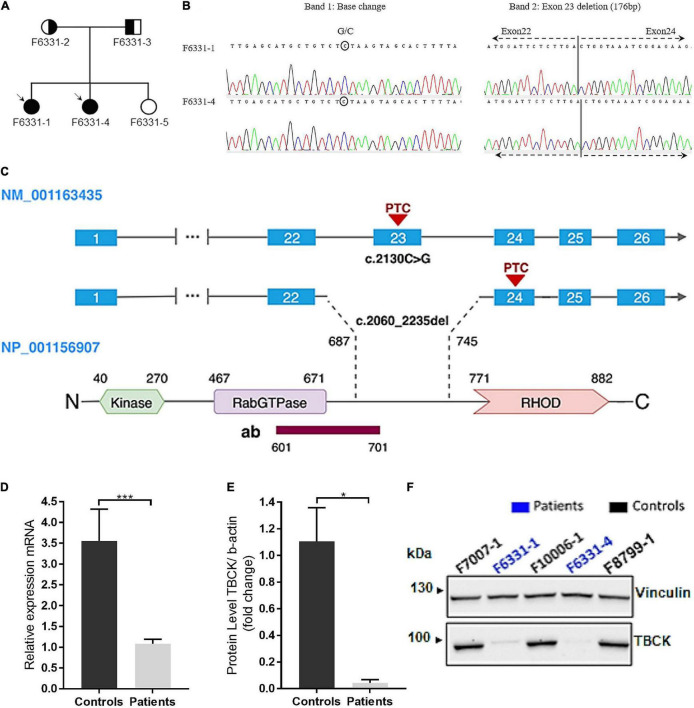
FIGURE 1.
IHPRF3 patients harboring biallelic pathogenic variants in TBCK and relative quantification of mRNA and protein expression levels in iNPC. (A) Pedigree of family F6331. Circle—female; square—male; symbol-filled—affected patients; symbol empty—not affected sister; circle half-filled—harbors the heterozygous microdeletion; square half-filled—harbors the heterozygous stopgain variant. (B) Chromatograms of the cDNA sequence show that both alleles are expressed: in the left image, a base change in Band 1 (expected base G changed to a C in reverse strand) represents the stopgain variant; and in the right image, the juxtaposition of exons 22 and 24 (excision of exon 23) in Band 2 representing the allele with the microdeletion. Bands 1 and 2 represent cDNA PCR products for TBCK in agarose gel (Supplementary Figure 2A). (C) Schematic representation of TBCK transcript and protein. The illustration shows the two mutant isoforms identified in the patients and the predicted positions on the protein. The antibody target region is also represented. The pathogenic variants create a putative premature termination codon (PTC) in exons 23 and 24, located within the C-terminal region of the protein, upstream of the RHOD domain. (D) TBCK mRNA expression (data from TBCK-mRNA-3 primer pair) shows reduced transcript levels in patients, compared with controls. Each biological sample had three technical replicates, and each individual was considered a biological replicate. (E,F) Western blot (WB) shows low levels of TBCK protein in iNPC of patients, compared with iNPC of controls, which are compatible with an autosomal recessive inheritance condition. *p-value < 0.05, ***p-value < 0.001, and n.s.—not significant. Data on graphs are shown as mean ± SD of technical replicates and biological samples. Patients: F6331-1 and F6331-4 (N = 2); controls: F7007-1, F8799-1, and F10006-1 (N = 4).