Abstract
Nuclear lamins are ancient type V intermediate filaments with diverse functions that include maintaining nuclear shape, mechanosignaling, tethering and stabilizing chromatin, regulating gene expression, and contributing to cell cycle progression. Despite these numerous roles, an outstanding question has been how lamins are regulated. Accumulating work indicates that a range of lamin post-translational modifications (PTMs) control their functions both in homeostatic cells and in disease states such as progeria, muscular dystrophy, and viral infection. Here, we review the current knowledge of the diverse types of PTMs that regulate lamins in a site-specific manner. We highlight methods that can be used to characterize lamina PTMs whose functions are currently unknown and provide a perspective on the future of the lamin PTM field.
Keywords: PTMs, acetylation, phosphorylation, ubiquitination, farnesylation, lamins
THE ROLES OF LAMINS IN NUCLEAR HEALTH AND DISEASE
Nuclear lamins (see Glossary) form a filamentous network adjacent to the inner nuclear membrane (Figure 1). These type V intermediate filaments, present in all multicellular metazoans, are thought to be the most ancient intermediate filaments [1–4]. Lamins, which are of type A (major isoforms A and C and minor isoforms AΔ10 and C2) or type B (lamins B1, B2, and B3), regulate fundamental nuclear processes (Box 1). Their functions include defining nuclear shape, mechanosignaling, participating in stress responses, aiding spatial organization within the nucleus, tethering and stabilizing chromatin, regulating gene expression, influencing DNA replication and repair, and contributing to cell cycle progression [5–12]. As expected for proteins with such diverse roles, mutations in nuclear lamins manifest in the development of a range of rare, but often autosomal dominant and devastating diseases, known as laminopathies. These include multiple diseases associated with lamin A (e.g. Emery-Dreifuss muscular dystrophy, Dunnigan-type familial partial lipodystrophy, dilated cardiomyopathy, and Hutchinson-Gilford progeria syndrome (HGPS)) [5]. HGPS, a well-studied lamina disease, stems from a point mutation that creates a cryptic splice site resulting in the deletion of 50 amino acids close to the C-terminus of lamin A [13,14]. This deletion removes the Zmpste24 cleavage site and induces the expression of a permanently farnesylated, dominant mutant of prelamin A known as progerin. Additional diseases, including autosomal dominant leukodystrophy [15] and acquired partial lipodystrophy [16], are associated with lamins B1 and B2, respectively. Lamin dysregulation also is linked to cancer biology. Lamin rearrangement maintains the balance between nuclear rigidity and pliability as cells migrate through small and stiff spaces during metastasis [17–19], and decreased lamin B1 expression induces poor prognostic outcomes for lung cancer [20].
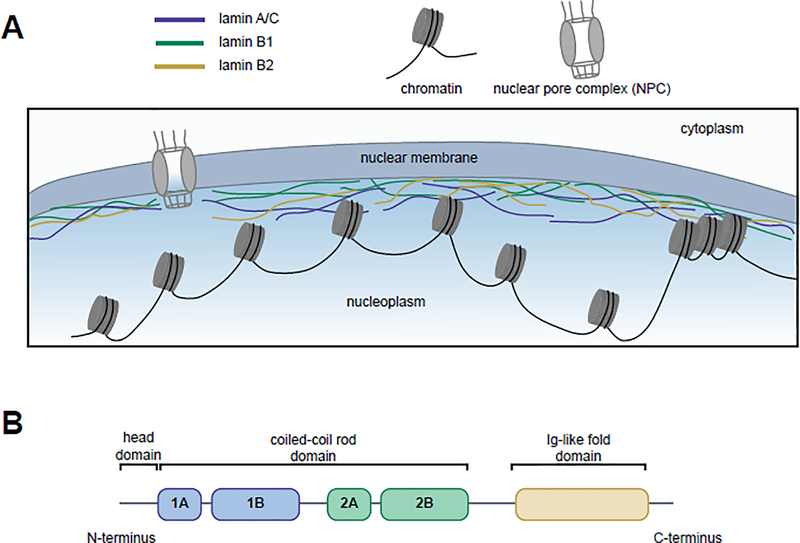
Figure 1. Lamins’ localization to the nuclear periphery and functional domains.
(A) The human lamins, lamin A/C (blue), lamin B1 (green), and lamin B2 (yellow), assemble into a network at the inner nuclear periphery where they serve to maintain nuclear shape and interact with both euchromatic and heterochromatic regions of DNA. (B) Lamins have three domains: a head domain, a coiled-coil rod domain (composed of four sub-domains) that mediates interactions with other lamina proteins, and an Ig-like fold domain that mediates interactions with non-lamina proteins.
Box 1. Lamins have distinct roles within the nucleus.
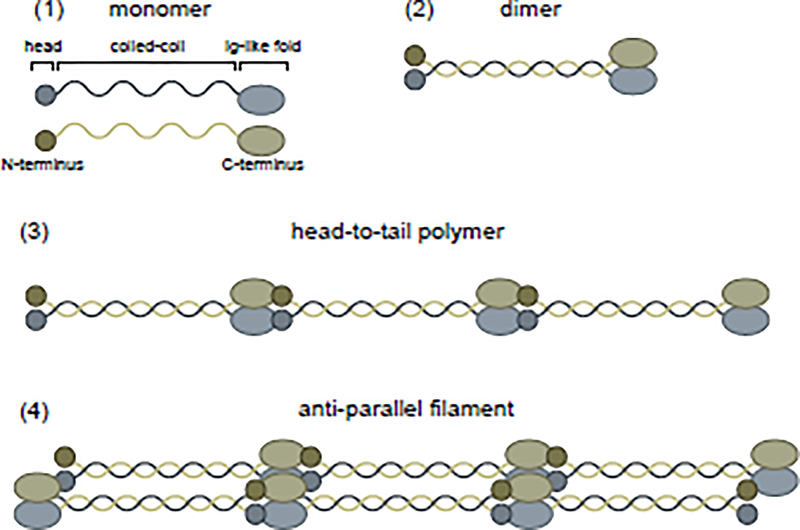
Lamins are divided into two subtypes: the separately encoded B-type lamins, lamin B1 and lamin B2, and the splice-variant-derived A-type lamins including two major (lamin A and lamin C) and two minor (AΔ10 and C2) isoforms [3,4,105,106]. All lamins share a set of domains: head, coiled-coil rod (Coil 1A, Coil 1B, Coil 2A, and Coil 2B), and globular Ig-like fold (see Figure 1B). To form a meshwork at the inner nuclear periphery, lamins assemble into higher order filaments by dimerizing via their α-helical coiled-coil rod domains (Figure I), forming head-to-tail polymers, and assembling in an antiparallel manner into filaments [107,108]. Lamins A/C, B1, and B2 form interacting but distinct networks, indicating likely lamin-specific functions [109–111]. This prediction is supported by reports that knockout or mutation of individual lamins results in different nuclear periphery morphological phenotypes [110–115]. The relative levels of each lamin at the nuclear periphery contribute to nuclear stiffness [116]. The ratio of A to B-type lamins dictates the degree to which the nucleus can be deformed without rupturing or damaging DNA during cellular migration through small spaces [17,117]. A-type lamins confer viscosity, helping to prevent rupture, while B-type lamins provide elasticity, enabling nuclear deformation [17]. Crystallographic and cryo-electron microscopy studies have shown that, despite the overall similarities in lamin structures [118,119], there are differences in the homodimer interfaces between lamin B1 and lamin A/C that contribute to their distinct networks [120–122].
Lamins also interact with heterochromatic (via lamin associated domains (LADs)) and euchromatic regions of DNA to help mediate genome organization, transcription, and DNA repair [11,78,123]. Lamin A/C was reported to serve as a scaffold for recruiting histone modifying enzymes, such as PCAF and HDAC2, to chromatin [124]. Beyond the nuclear periphery lamins, a subset of A-type lamins in the nucleoplasm also is linked to transcriptional regulation [76]. Additionally, the association between lamins and DNA has been implicated in determining DNA repair pathway choice (nonhomologous end joining (NHEJ) or homologous recombination (HR)) [12,125]. In line with A and B-type lamins having distinct roles, the lamin A network is dissolved early in mitosis, while lamin B1 remains associated with the fragmented nuclear membrane [45]. Furthermore, lamin B1 localizes to daughter nuclear membranes earlier than lamin A [126,127]. These findings suggest that lamin B1 may facilitate re-formation of the nuclear periphery upon completion of mitosis.
Figure I. Lamins assemble into filaments.
Lamins, initially synthesized as monomers (1), assemble into intermediate filaments by first dimerizing (2) and then forming head-to-tail polymers (3). These polymers assemble in an anti-parallel manner into filaments (4).
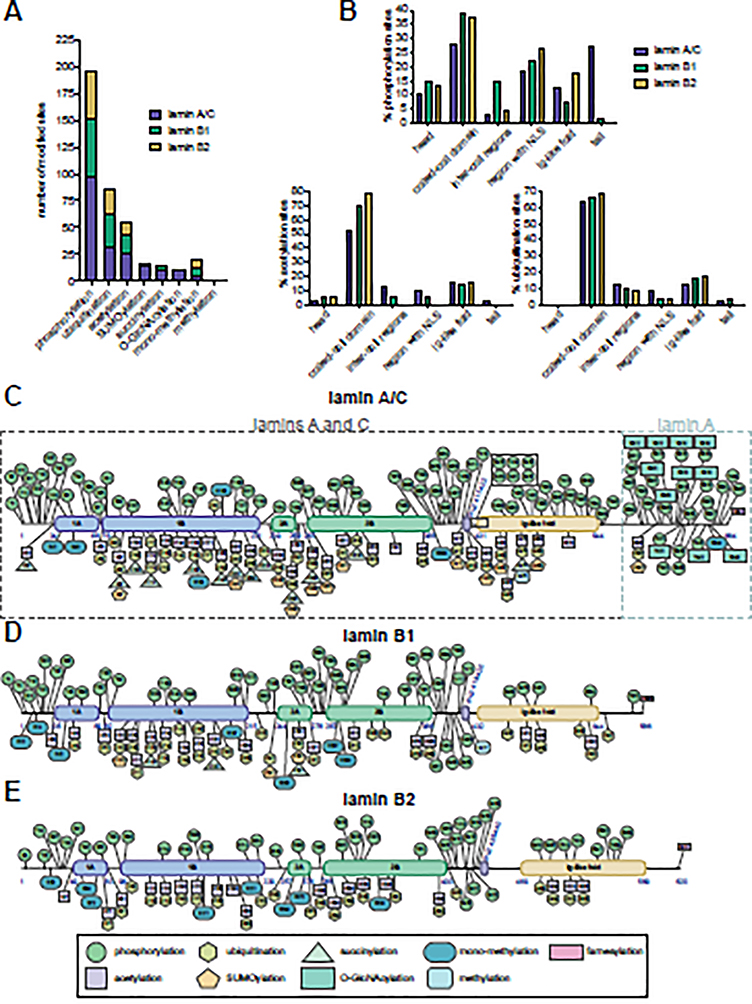
An underlying challenge in lamin biology is determining how lamins are regulated in homeostatic and disease states. While recent work has highlighted that protein-protein, protein-DNA, and protein-RNA interactions contribute to the multifaceted functions of lamins [10,21–25], accumulating evidence shows that nuanced lamin functions are regulated through post-translational modifications (PTMs). Advances in mass spectrometry (MS) have facilitated the identification of diverse PTMs on lamins, including acetylation [26–28], farnesylation [29], methylation [30], arginine monomethylation [31], O-GlcNAcylation [32–34], phosphorylation [35–37], succinylation [38], SUMOylation [39], and ubiquitination [40,41] (Figure 2A). Some of these PTMs, such as farnesylation, occur during post-translational processing [42,43], while others occur under specific biological conditions. In addition, the number of sites modified per lamin, and their distribution across lamin domains, varies. For example, phosphorylation was detected across the domains (Figures 1B, 2B–E), while acetylation and ubiquitination have been primarily found within the coiled-coil rod domain (Figures 1B, 2B–E). While the roles for many of these site-specific PTMs remain unknown, significant progress has underlined their impact on lamin biology. Here, we discuss the complex functions of lamin PTMs, implications in human disease, methods for PTM detection and characterization, and future considerations for the growing lamin PTM field.
Figure 2. The expanding landscape of human lamin post-translational modifications.
(A) Number of modifications on the human lamins per PTM type. (B) Percentage of phosphorylation (upper), acetylation (lower left), and ubiquitination (lower right) sites in the different lamin domains. (C-E) Human lamin PTMs documented on PhosphoSitePlus and/or discussed in this review are indicated for (C) lamin A/C (amino acids shared between lamins A and C and those unique to lamin A are indicated by dashed boxes), (D) lamin B1, and (E) lamin B2. The first and last amino acids of the different domains and the amino acids for the nuclear localization signal (NLS) are indicated in blue.
LAMIN DYNAMICS AND NUCLEAR SHAPE
A function of PTMs that is becoming well-established is the regulation of lamin dynamics that informs nuclear shape. The PTM-mediated regulation of lamin integrity and arrangement at the nuclear periphery is fundamental to cellular homeostasis, and its misregulation is linked to several disease phenotypes.
PTMs regulate lamins during mitosis
The first dynamic lamin PTM to be detected over 30 years ago was phosphorylation during mitosis. Early in mitosis, the nuclear lamina is disrupted to enable microtubules to extend into the nuclear space and attach to chromosomes. A key regulator of this disruption is phosphorylation. All three human lamins have a conserved phosphorylation site in the head domain that is required for disassembly during mitosis (lamin A/C S22, lamin B1 S23, lamin B2 S37) [9,44–46] (Figure 3Ai). Lamin A/C S392 phosphorylation is also important for mitotic nuclear envelope breakdown. While multiple kinases can modify these sites, CDK1 and PKC are most frequently reported [9,47,48]. Additional lamin A sites also are phosphorylated by CDK1 (S392) and PKC (S5, T199, T416, T480, S625) in what is likely a temporal regulation of lamin disruption during cell cycle progression [49]. Similar to lamin A/C, lamin B1 phosphorylation at S395, S405, and S408 by PKC leads to lamina and nuclear envelope disruption [50]. Upon completion of mitosis, removal of lamin B phosphorylation by type 1 protein phosphatase (PP1) facilitates lamina reassembly [51] (Figure 3Ai).
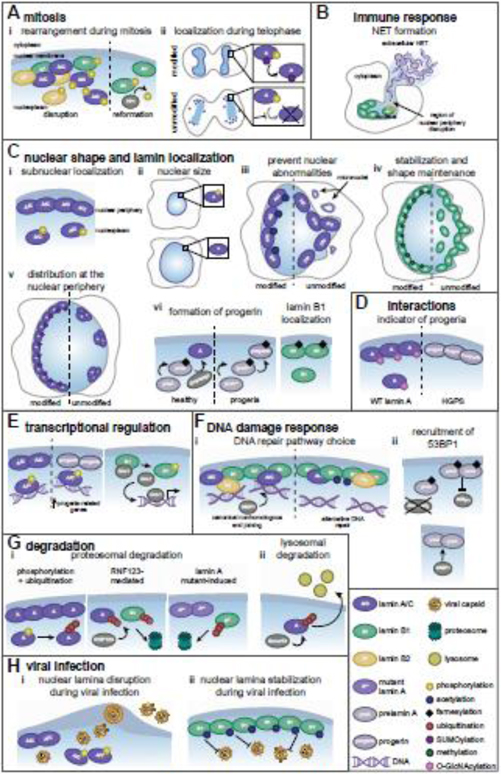
Figure 3. Post-translational modifications regulate lamin functions.
Mature lamin A/C (A/C), lamin A (A), lamin B1 (B1), lamin B2 (B2). (A) (i) During mitosis, lamin phosphorylation promotes nuclear periphery disruption. PP1-mediated B1 dephosphorylation facilitates lamina reformation after mitosis. (ii) At telophase, SUMOylation promotes A dephosphorylation and nuclear relocalization. (B) B1 phosphorylation disrupts the nuclear periphery, releasing chromatin for NET formation by neutrophils. (C) (i) A/C phosphorylation determines sub-nuclear localization. (ii) A phosphorylation maintains nuclear size. (iii) A/C acetylation prevents nuclear deformations. (iv) B1 methylation maintains its localization and nuclear shape. (v) A SUMOylation maintains lamin spacing. (vi) A maturation requires Zmpste24-mediated cleavage of prelamin A and farnesylation removal, which are absent in progeria. Farnesylation promotes proper B1 localization. (D) O-GlcNAcylation decorates wild type A but not progerin. (E) A/C phosphorylation enables its association with enhancers, which is altered by progerin. B1 phosphorylation releases Oct-1. (F) (i) In homeostasis, B1 acetylation increases and decreases associations with lamins and chromatin, respectively. Upon DNA damage, this reduced chromatin association modulates selection of DNA repair pathways. (ii) Farnesylation status of prelamin A impacts 53BP1 recruitment to damaged DNA. (G) (i) A phosphorylation promotes its ubiquitin-mediated degradation, and RNF123-mediated A and B1 ubiquitination induces their degradation. A mutants present in Emery-Dreifuss muscular dystrophy promote B1 ubiquitination-mediated degradation. (ii) Smurf2-mediated ubiquitination is implicated in lysosome-mediated A degradation. (H) (i) A/C phosphorylation during viral infection induces lamina disruption, facilitating viral capsid nuclear egress. (ii) During herpesvirus infection, B1 acetylation inhibits lamina disruption and viral capsid egress.
More recently, it has been shown that the interplay between SUMOylation and phosphorylation contributes to lamina reformation [52]. Specifically, a SUMO site (residues 494–497) located in the lamin A Ig-like fold domain influences the accumulation of lamin A on chromosomes during telophase [52] (Figure 3Aii). When this SUMOylation is blocked, dephosphorylation of S22 is reduced, and the subsequent nuclear reorganization after mitosis is impaired. This finding underscores the importance of crosstalk between these PTMs to coordinate lamin dynamics during mitosis. The balance between well-characterized and less studied lamin PTMs likely will provide insight into complex lamin dynamics.
Lamin PTMs in immune response
Dissolution of the lamina, initially characterized as a mitotic event, has subsequently been shown to function in other cellular processes. Specifically, during the inflammatory response, effective lamina breakdown is needed to generate neutrophil extracellular traps (NETs) [53]. NETs are a mixture of chromatin and proteins that are extruded from the cell to ensnare pathogens. The ability of this chromatin to serve as the scaffold for NETs is dependent on the externalization of chromatin from the nucleus via the disassembly of the nuclear lamina (Figure 3B). Phosphorylation of lamin B1 at S395, S405, and S408 by PKC facilitates lamina disruption and allows the release of chromatin for NET formation [53]. Inhibition of PKC, mutation of these lamin B1 sites to prevent their phosphorylation, or lamin B1 overexpression all result in decreased lamina disruption and NET release. Underscoring the relevance of NETs to human disease, a recent study of SARS-CoV2 patients showed that sustained levels of NETs in the blood of intensive care patients correlated with heightened mortality [54]. This finding suggests that NETs, and the perturbation of the lamina that proceeds their formation, may be a future therapeutic target.
Nuclear shape and lamin localization
PTM-mediated regulation of lamin organization and localization is a key factor in maintaining nuclear shape and integrity in interphase cells. Lamin A/C phosphorylation contributes to nuclear size and lamin sub-nuclear localization (Figure 3Ci). In a phosphoproteomic study, Kochin, et al. investigated lamin A phosphorylation during interphase, identifying twenty sites in three regions: N-terminus (7 sites), between the rod domains and the Ig-fold (10 sites), and far C-terminus (3 sites) [55]. Eight phosphorylation sites were characterized as high-turnover sites, including S22 and S392. Using point mutations, phosphorylation at these sites was shown to dictate the ratio of nuclear periphery versus nucleoplasmic subsets of lamin A, with S22 having the dominant effect [55]. Beyond mediating subnuclear localization, lamin phosphorylation also contributes to nuclear localization. PKC-mediated phosphorylation at lamin A S403 and S404, located close to the nuclear localization signal (NLS), facilitates lamin A nuclear import [56]. Whether other PTMs, such as NLS-localized K417 methylation of lamin B1 [30], also impact lamin subcellular localization remains to be determined.
Lamin phosphorylation also mediates nuclear shape and stability. For example, phosphorylation of lamin A S268, a site conserved across species, reduces nuclear size [57]. Mutation of this site to an unmodifiable amino acid induces nuclear enlargement in a manner reminiscent of the alterations in nuclear size observed in cancer (Figure 3Cii). Additionally, AKT-mediated modification of mature lamin A S404 contributes to nuclear stability [58], and S404 mutation induces nuclear abnormalities similar to those in Emery-Dreifuss muscular dystrophy [59]. More broadly, lamins function in cellular mechanosensing through their connection across the nuclear envelope to the cytoskeleton. Studies have connected lamin A/C S22 phosphodynamics to substratum stiffness [60]. Increased substratum stiffness induces increased cell spreading and cytoskeletal tension, which lead to decreased S22 phosphorylation and increased nuclear stiffness. This dephosphorylation correlates with increased myosin-IIA expression [60]. This connection between S22 phosphodynamics and nuclear mechanics may influence nuclear stability during metastasis, during which cells squeeze through constrictive spaces [17]. Additionally, phosphorylation events identified as disease-specific may also be linked to the regulation of lamin integrity. Phosphorylation by AKT1 of S458 in A-type lamins has only been detected in myopathies caused by mutations in lamin A [61]. NMR and CD spectra demonstrated that the R453W mutation, which occurs in Emery-Dreifuss muscular dystrophy, destabilizes the lamin A structure [62]. It is predicted that this lamin unfolding makes S458 accessible for phosphorylation; however, it remains unknown whether this modification is a mechanism to counteract or enhance the disease phenotype.
Recent studies have illustrated how other lamin PTMs, including acetylation, methylation, and SUMOylation, are also important for maintaining lamin localization and nuclear periphery integrity. Acetylation of lamin A/C K311 is needed to prevent nuclear blebbing and other nuclear abnormalities, including the formation of micronuclei [63] (Figure 3Ciii). Karoutas, et. al. determined that the acetyltransferase MOF/MYST1/KAT8 is responsible for acetylating lamin A/C K311. Without this acetylation, nuclear integrity and genome organization are compromised. Supporting the importance of this modification, Murray, et al. also observed persistent K311 acetylation of lamin A/C [64]. Methylation by EHMT1 and EHMT2 (K417) also can stabilize lamin B1 K417 [30] (Figure 3Civ). Mutation of this lysine to an alanine resulted in lamin B1 nucleoplasmic retention and decreased nuclear periphery localization. When EHMTs were silenced, the nuclear periphery was deformed. Additionally, it is becoming increasingly apparent that SUMOylation of lamins also impacts their dynamics and is connected to laminopathies. Both SUMO1 and SUMO2 act on lamin A, and K201 can be SUMOylated [65]. When the disease-inducing mutants E203G, E203K, and K201R are present in familial dilated cardiomyopathy, there is reduced K201 SUMOylation [65]. This alteration in PTM status results in a punctate, irregularly spaced array of lamin A (instead of a smooth lamin A ring) at the nuclear periphery [65] (Figure 3Cv) and leads to increased cell death. The elevated cell death induced by the E203K mutation may point to a mechanism by which SUMOylation promotes nuclear health.
Another PTM that has long been recognized as a fundamental contributor to lamin functions is farnesylation during lamin protein maturation. Both the B-type lamins and prelamin A, but not lamin C, have a C-terminal CaaX motif in which the cysteine is farnesylated [29,42,43,66] (Figure 2C–E). During processing of prelamin A into mature lamin A, the C-terminal 15 amino acids, including the farnesylated cysteine, are cleaved by Zmpste24 [43,67,68] (Figure 3Cvi). In HGPS, the Zmpste24 cleavage site is absent. This omission leads to expression of the disease-inducing farnesylated version of prelamin A known as progerin [13,14]. In contrast to lamin A, lamin B1 is farnesylated in its mature form [29]. This PTM is a lipid anchor that facilitates the close and uniform association between lamin B1 and the nuclear membrane [69] (Figure 3Cvi). If lamin B1 is not farnesylated, it does not properly localize to the nuclear periphery [42], and this mislocalization has detrimental consequences. For example, if nonfarnesylated lamin B1 is expressed in developing mice, they die soon after birth due to severe neurodevelopmental defects caused by nuclear abnormalities in which the chromatin detaches from the nuclear periphery [69]. Thus, proper lamin farnesylation status is essential for maintaining lamin functions.
Emerging lamin PTMs and protein-protein interactions
Two less studied PTMs, SUMOylation and O-GlcNAcylation, are connected to disease states and predicted to alter lamin protein-protein interactions. In familial partial lipodystrophy, lamin A mutations G465D and K486N occur in a SUMO motif located in the Ig-like fold domain, a region that mediates lamin protein-protein interactions [70]. In the presence of these mutations, SUMOylation is impaired, which likely affects protein associations with this domain [70]. Multiple studies have detected lamin A O-GlcNAcylation, which is the addition of the sugar ß-O-linked N-acetylglucosamine by O-GlcNAc transferase (OGT) [32–34,71]. This glycosylation localizes towards the C-terminus (amino acids 385–646), and contributes to a “sweet spot” (amino acids 601–645) of eleven O-GlcNAcylation sites [34] (Figure 2C). This area overlaps with the region that is required for Zmpste24-mediated prelamin A cleavage, which means that, in HGPS, part of this region is deleted. O-GlcNAcylation was not detected in the presence of the HGPS mutant (Figure 3D). It is likely that the amino acids that are deleted in this disease are required for either substrate recognition and/or direct modification of lamin A by OGT [34]. While crosstalk between O-GlcNAcylation and phosphorylation has not been determined for lamin A, crosstalk between these two PTMs has been found for multiple cell cycle progression proteins and contributes to cell division pathways [32]. Based on these findings and the knowledge that the same lamin A residues have been identified as O-GlcNAcylated or phosphorylated (Figure 2C), it is likely that the interplay between these PTMs contributes to regulating lamin A during mitosis and nuclear envelope reassembly [32]. In addition, acetylation is a putative regulator of lamin-protein interactions. Several acetyltransferases and deacetylases, including HDAC2, HDAC3, and SIRT6, are known to localize to the nuclear periphery and interact with nuclear periphery proteins or lamins [72–75]. Notably, association between lamin A/C and HDAC2 was lost in Emery–Dreifuss muscular dystrophy myoblasts expressing mutant lamin A [74].
Altogether, both well-studied PTMs, such as phosphorylation and farnesylation, and emerging lamin PTMs, such as acetylation, methylation, SUMOylation, and O-GlcNAcylation, contribute to lamin dynamics and nuclear shape, and are critical in both homeostatic and disease states.
TRANSCRIPTIONAL REGULATION
Accumulating reports have also linked lamin phosphorylation to transcriptional regulation. The phosphorylation of S22 and S392 that induces nucleoplasmic lamin A/C also enables its association with kilobase-wide genomic enhancer binding sites near active genes [76] (Figure 3E). Since phosphomimetic lamin C, but not lamin A, bound enhancers that also were bound by WT lamin A/C, this association may point to a previously unrecognized specific function for lamin C. Phosphorylated lamin A/C bound different enhancers in cells expressing progerin than in normal cells. In the presence of progerin, the DNA regions that are differentially bound cause upregulation of genes that are clinically relevant to progeria progression [76] (Figure 3E). While progerin itself is not phosphorylated at S22 [76], it is likely that the interaction between progerin and mature lamin A/C induces a misdirection of phosphorylated lamin A/C to different enhancers and causes the subsequent expression of genes related to progeria. Lamin phosphorylation also contributes to targeted alterations in gene expression [60,77]. For example, JNK-mediated phosphorylation of lamin B1 T575 releases the transcription factor Oct-1 during genotoxic stress, inducing expression of Oct-1 target genes [77] (Figure 3E). As lamins have been shown to interact with chromatin [10,11,78], it is likely that future work will uncover additional examples of biological context-dependent lamin-mediated transcriptional regulation.
DNA DAMAGE RESPONSE
It is becoming well-understood that lamins function in the DNA damage response, and lamin PTMs, including acetylation, farnesylation, and SUMOylation, contribute to DNA repair pathways. Recent work has established that lamin B1 K134 acetylation decreases the association between lamin B1 and chromatin and impedes cell cycle progression upon DNA damage [79] (Figure 3Fi). Acetylation at this site inhibits DNA repair by canonical nonhomologous end joining [79], which depends on tethering chromatin to the nuclear periphery [12]. Increased levels of K134 acetylation were observed during the S/G2 phases of the cell cycle when homologous recombination [79], which does not require tethering of chromatin to the lamina, would be the preferred pathway to repair damaged DNA. Thus, by mediating the association between the nuclear periphery and chromatin, this lamin B1 acetylation likely acts as a molecular toggle in DNA repair pathway choice in a cell cycle stage-dependent manner [79]. It also has been shown that the lamin A/C and HDAC2 interaction is reduced after oxidative stress [75]. HDAC inhibitors strengthen this association, while drug-induced accumulation of prelamin A weakens this association [80]. These findings suggest that the interaction between lamins and HDACs may be dependent on HDAC activity. Whether this interaction leads to HDAC-mediated deacetylation of lamins remains to be determined.
Lamin A farnesylation status also is connected to DNA repair [81]. When Zmpste24 is knocked out and farnesylated prelamin A cannot be processed, DNA repair is delayed due to defective 53BP1 recruitment [82]. This is likely because farnesylated prelamin A has an aberrant tight association with the nuclear lamina, thereby being less mobile to mediate DNA repair (Figure 3Fii). Lattanzi, et al. showed that expression of non-farnesylated prelamin A induces 53BP1 recruitment and promotes DNA repair after oxidative stress [83] (Figure 3Fii). Thus, the farnesylation status of prelamin A as a regulator of DNA repair merits future investigation.
Upon extensive DNA damage, lamin A/C SUMOylation promotes its association with LC3, a protein associated with the nucleophagy, autophagy, and lysosomal degradation pathways [84]. LC3 also associates with lamin B1 upon DNA damage [85]. This interaction suggests that the SUMOylation-mediated interaction between lamins and LC3 may be a mechanism to promote cell death in cases of extensive DNA damage that could lead to tumorigenic conditions.
LAMIN DEGRADATION
The degradation of lamins is an important process that can be beneficial or detrimental to a cell depending on biological context, and phosphorylation and ubiquitination have been found to contribute to this regulatory mechanism. For example, in a process predicted to conserve resources, AKT phosphorylates prelamin A S404 in G2/M phases of the cell cycle. This event triggers prelamin A redistribution to the cytoplasm and lysosomal degradation [58,86]. In this case, phosphorylation likely induces clearance of excess prelamin A before commitment to its multi-step post-processing maturation. Phosphorylation also impacts lamina disruption during apoptosis. Specifically, PKC-mediated lamin B S395 and S405 phosphorylation induces lamin B solubilization, followed by lamina disassembly and lamin B degradation. Together, these events facilitate DNA fragmentation and apoptosis [87]. Similarly, lamin A/C S392 CDK5-mediated phosphorylation induces lamin A/C dissolution, which enables its ubiquitination and subsequent degradation [88] (Figure 3Gi). The type VI intermediate filament nestin can translocate to the nucleus and protect lamin A/C from CDK5-induced degradation by suppressing CDK5 enzymatic activity [88]. This inhibition of lamin A/C degradation helps to maintain nuclear integrity and protects tumor cells from becoming senescent [88]. It is possible that similar mechanisms protect the B-type lamins from degradation.
Ubiquitination was shown to facilitate the proteasomal degradation of lamins [89,90]. Several ubiquitin ligases target lamins, including RNF123, HECW2, Smurf2, and Siah1 [89–92]. Some of these interactions have been studied in more detail. Both lamin B1 and lamin A have been identified as substrates of RNF123 [90] (Figure 3Gi). Interestingly, lamin B1 is targeted for proteasomal degradation upon expression of the lamin A point mutants G232E, Q294P, and R386K, mutations that are present in Emery-Dreifuss muscular dystrophy [90] (Figure 3Gi). Such investigations highlight the broad impact of lamin A point mutations on nuclear architecture and functionality in disease states. Another enzyme upregulated in Emery-Dreifuss muscular dystrophy, HECW2, was also shown to ubiquitinate lamin B1, targeting it for proteasomal degradation [89]. Lamin ubiquitination also is linked to the autophagic-lysosomal pathway. Smurf2-mediated ubiquitination of lamin A induces its lysosomal, but not proteasomal, degradation [91] (Figure 3Gii). Future work is needed to tease apart the site-specificity of how these distinct ubiquitin ligases mediate lamin function and turnover during both homeostatic and disease states.
VIRAL INFECTION
Lamin PTMs are implicated in mediating lamina stability during viral infection. The same phosphorylation site that is critical for disruption of lamin A/C during mitosis is repurposed by herpesviruses to facilitate viral capsid nuclear egress. These viruses replicate and package their genomes into viral capsids in the host nucleus. However, once packaged, the viral capsids (~125 nm) are too large to pass through a nuclear pore. Consequently, these viruses must disrupt the lamina so that viral capsids can bud through the nuclear membrane to reach the cytoplasm and complete their assembly (Figure 3Hi). Herpes simplex virus type 1 and type 2 (HSV-1, HSV-2, alphaherpesviruses), human cytomegalovirus (HCMV, betaherpesvirus), and Epstein-Barr virus (EBV, gammaherpesvirus) all encode a viral kinase that phosphorylates lamin A/C S22 to facilitate localized lamina disruption [93–98]. Other nuclear-replicating viruses, including baculovirus and porcine circovirus type 2, rely on localized lamina disruption via S22 phosphorylation for effective virus production [99,100]. Increasing evidence suggests that these viruses may be co-opting a host pathway that enables transport of large protein complexes across the nuclear membrane. Specifically, Speese, et al. showed that, in Drosophila, ribonucleoprotein particles (RNPs), which are too large to fit through a nuclear pore, bud into the nuclear membrane upon A-type lamin disruption induced by atypical PKC-mediated phosphorylation [101]. This budding facilitates RNP granule nuclear export. Thus, lamin A phosphorylation by viral kinases that enables transit of viral capsids across the nuclear membrane likely reflects a repurposing of this endogenous pathway for nuclear egress of large complexes.
Recent work has shown that lamin acetylation may play a previously unrecognized antiviral role during herpesvirus infection. While studying the impact of the human pathogen HCMV on the cellular acetylation landscape, Murray, et al. demonstrated the temporal modulation of lamin acetylation at lamin A/C (19 sites), lamin B1 (8 sites), and lamin B2 (10 sites) during infection [64]. Increased lamin B1 acetylation was observed at late stages of viral infection. Molecular virology and microscopy assays uncovered that lamin B1 K134 acetylation stabilizes the nuclear lamina during HCMV infection and inhibits virally-induced nuclear curvature and nuclear periphery disruption (Figure 3Hii). This increased nuclear integrity causes an accumulation of viral capsids within the nucleus and results in reduced infectious virus production [64]. Additional work has demonstrated that K134 acetylation also maintains nuclear integrity during HSV-1 infection [79]. As the analogous amino acid residue in lamin A/C and lamin B2 is an arginine, it is likely that K134 acetylation acts as a specific regulator of lamin B1. Together, these studies of phosphorylation and acetylation during viral infection underscore the importance of future work on the crosstalk between lamin PTMs that may occur during the same biological process.
Altogether, it is clear that substantial progress has been made in uncovering the range of PTMs that decorate lamins. This progress was facilitated by technological developments, including MS-based methods, for identifying and characterizing lamin PTMs (Box 2). With this expanding knowledge of lamin PTMs comes the challenge of understanding their temporal and spatial regulatory functions, their interconnectivity, and their contribution to homeostatic cellular processes and disease states.
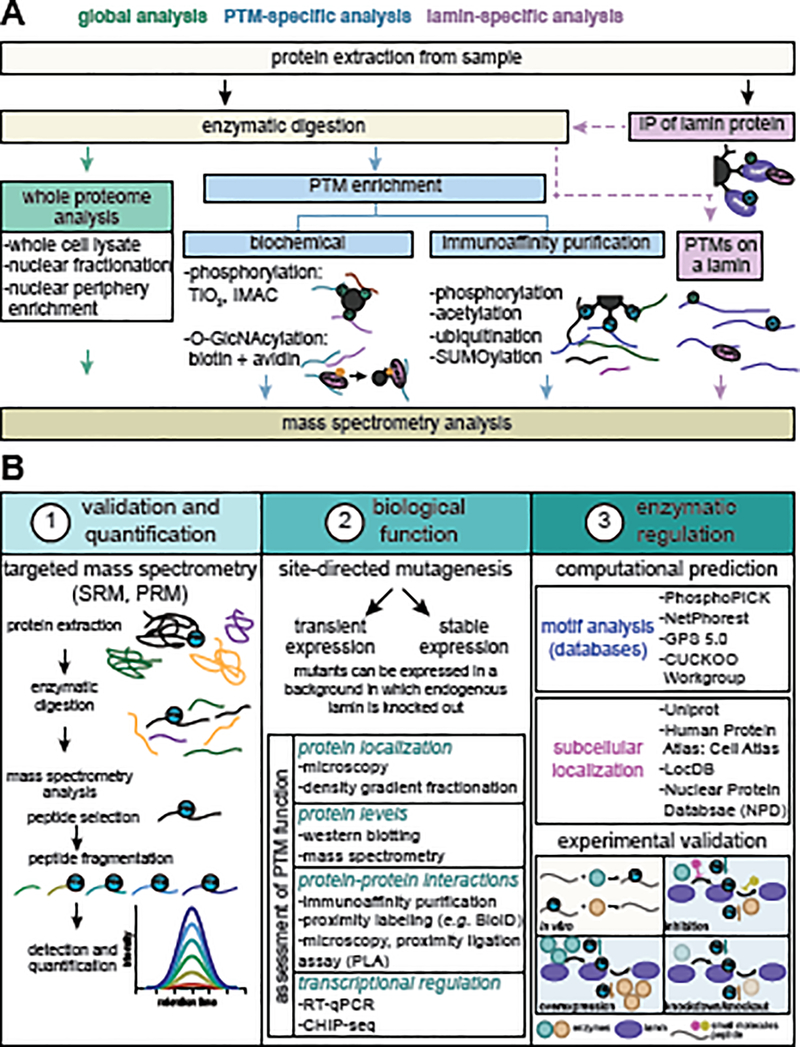
Box 2. Methods to detect and characterize lamin PTMs.
Technology advancements in post-translational modification (PTM) enrichment methods and mass spectrometry (MS) have expanded the ability to detect lamin PTMs (Figure IA). Proteomic workflows, including TiO2 phosphopeptide enrichment, have confirmed the lamin S22 and S392 phosphorylations (originally detected by 32P-labeling, two-dimensional thin-layer chromatography and electrophoresis, and reverse phase chromatography [46]) and expanded the knowledge of lamin phosphorylations [55,128,129]. PTM-specific antibodies suitable for immunoaffinity purifications and employment of different enzymatic digestions have enhanced the MS-identification of diverse lamin PTMs, including acetylation [26,64,130], ubiquitination [40,41,131], and SUMOylation [39,132]. Chemoenzymatic approaches that use biotinylation plus avidin-mediated capture have identified O-GlcNAcylated lamin A sites [32,33]. These advances have paved the way for understanding the scope and biological functions of the lamin PTM landscape.
Upon PTM identification, the next steps are (1) validating and quantifying the modification, (2) determining its biological function, and (3) understanding its regulation (Figure IB). Targeted MS (selected or parallel reaction monitoring) offers an accurate method for site-specific PTM confirmation and quantification. Targeted MS detects modified peptide(s) of interest using signature parameters, which are transferrable and broadly applicable across labs and biological conditions. While not yet used for lamin PTMs, recent studies have demonstrated its value for characterizing several PTMs [133–137].
To interrogate the function of a particular lamin PTM, classical methods use site-directed mutagenesis to mimic either modified or unmodified versions of the residue of interest. Generation of cells expressing these mimic constructs usually relies on transient or stable expression. CRISPR/Cas9 technology enables construct expression in a knockout background [79], as well as knockin strategies for expressing constructs from endogenous promoters. Lamin mutants can be probed using functional assays [9,30,53,63,64,79].
To understand how lamin PTMs are regulated, one considers possible enzymatic or non-enzymatic mechanisms. So far, lamin PTMs have only been shown to occur enzymatically. Databases, including GPS 5.0, PhosphoPICK, and NetPhorest 2.1 [138,139], can predict the kinases/phosphatases that may regulate phosphorylation. The GPS Cuckoo Workgroup is a resource for predicting regulatory enzymes. As lamins typically localize to the nuclear periphery, additional predictions can derive from the enzyme’s subcellular localization. Such predictions enable in vitro, tissue culture, and in vivo experiments to validate the enzymatic site-specific activity. Drug-targeting of the enzyme, CRISPR/Cas9-mediated knockouts, or shRNA/siRNA-mediated knockdowns can assess this site-specific activity [26,63]. Ongoing work to define lamin PTM levels, stoichiometry, function, and regulation of will continue to expand the understanding of lamin biology.
Figure II. Methods to detect and functionally characterize lamin PTMs.
(A) Lamin PTMs can be identified by MS using several experimental workflows, including whole proteome analyses (green), PTM-specific enrichment by biochemical or immunoaffinity purification methods (blue), or lamin-specific enrichment by immunoaffinity purification (IP) of a particular lamin (purple). (B) Considerations when characterizing lamin PTMs are (1) validation and quantification, (2) determination of biological function, and (3) assessment of enzymatic regulation. Targeted MS, including selected reaction monitoring (SRM) and parallel reaction monitoring (PRM), provides the means for confirming and accurately quantifying the levels of a site-specific PTM. Biological function can be determined by employing site-directed mutagenesis of the lamin of interest combined with functional assays that test different aspects of lamin biology (examples of functions and experimental techniques are listed). As many enzymes may regulate the same PTM, employing motif and subcellular localization computational analyses (examples of databases/tools are listed) can narrow the focus on which enzyme(s) may work on a particular PTM. The ability of the enzyme(s) to modify that site can then be tested using in vitro (tan) and cell culture and in vivo experiments (blue). Examples of the consequences on PTM levels of an enzyme that adds (teal) or removes (orange) a given PTM are shown.
LAMIN PTMs AND THERAPEUTIC DEVELOPMENTS
Due to the connection between lamin regulation and multiple disease states, therapeutic interventions that aim to alter lamin PTMs have become attractive for drug development. One example is the development of therapies against herpesviruses. These nuclear-replicating viruses remain latent throughout the lifespan of an infected individual. Since a large percentage of the adult population is already infected with these viruses, therapeutic interventions have included efforts to impede virus reactivation or, if a virus is reactivated, to suppress its nuclear egress. A promising antiviral drug currently in stage III clinical trials, maribavir, inhibits HCMV by targeting the virally-encoded kinase pUL97 [102,103]. Inhibition of this enzyme mitigates its ability to phosphorylate lamin A/C, thereby suppressing virus-induced nuclear envelope disruption and viral capsid nuclear egress. Maribavir is effective against HCMV strains that are resistant to the current antiviral treatment, ganciclovir, which targets the viral polymerase [104].
As nuclear periphery stability is often altered in laminopathies, druggable targets that can maintain or toggle nuclear stability may be beneficial. Acetylation and phosphorylation sites on lamins should be amenable to such therapeutic intervention, especially if the enzyme responsible for mediating the modification is known. Since MOF maintains the stabilizing lamin A/C K311 acetylation [63], this acetyltransferase may provide a target for increasing nuclear periphery mechanostability. As the pliability of the nucleus is important for cancer cell metastasis, identifying lamin PTMs that can be targeted to increase nuclear rigidity could provide an avenue for cancer treatment. In addition, some PTMs may serve as diagnostic tools for certain diseases. For example, lamin A/C O-GlcNAcylation is not detected in HGPS [34]. Assessment of the presence or absence of this modification could provide an additional method for diagnosing HGPS.
CONCLUDING REMARKS
This is an exciting time in understanding lamin biology. The identification of a diverse range of PTMs on lamin proteins has revolutionized our ability to understand their regulation in both biological and clinical contexts. It is now evident that dynamic lamin PTMs provide a critical hub for regulating nuclear integrity and the response to stimuli during various processes, including cell cycle progression, DNA damage response, immune signaling, mechano-responses, and pathogen infections. Given the ever-expanding view of the lamin PTM landscape, many questions remain unanswered about the roles of site-specific lamin PTMs (see Outstanding Questions). Adding to this challenge is the observation that certain PTMs, such as acetylation and ubiquitination, can occur on the same residue. How the exchange between these PTMs is regulated remains to be determined. Crosstalks also can also exist between PTM sites on the same lamin or different lamins to dynamically regulate the nuclear lamina meshwork in space and time across different cellular conditions. Targeted MS can help to define the stoichiometry of these PTMs, while follow-up in vitro and in cell culture functional analyses can further uncover PTM crosstalks. Understanding how PTMs provide layers of lamin regulation will not only help to inform fundamental nuclear biology, but also reveal how these regulatory mechanisms are disrupted in human disease states as diverse as genetic disorders, cancer progression, neurological disorders, cardiac disease, and virus-induced pathologies.
Highlights.
Lamins are ancient intermediate filaments that mediate critical nuclear processes by maintaining nuclear shape, stabilizing chromatin, regulating transcription, and contributing to cell cycle progression.
The two classes of lamins form overlapping networks at the nuclear periphery but have distinct regulatory roles.
Technological advances revealed an expansive range of post-translational modifications that decorate lamins, including phosphorylation, acetylation, ubiquitination, SUMOylation, methylation, and O-GlnAcylation.
While phosphorylation is a known regulator of mitotic laminar disruption, diverse post-translational modifications are being established as toggles of lamin arrangement, interactions, and functions.
The emerging field of lamin post-translational modification has wide-ranging implications in laminopathies, cancer progression, and viral pathogen infections.
What is the site-specific biological function of the diverse lamin PTMs that have been identified but not yet characterized?
What enzymatic or nonenzymatic mechanisms regulate distinct lamin PTMs and in what biological contexts?
How does lamin phosphorylation contribute to alterations in gene expression observed in aging, pathogen infections, and both hereditary and developed diseases?
To what extent do multiple PTMs that have been detected to modify the same site (e.g. acetylation, ubiquitination, and SUMOylation; O-GlcNAcylation and phosphorylation) crosstalk with each other?
How do PTMs on disctinct sites of the same lamin protein or on different lamins crosstalk with each other to induce global changes in laminar architecture? What determines which PTM(s) is dominant in determining lamin function?
How do these PTMs impact lamin-protein, lamin-DNA, and lamin-RNA interactions?
Are lamin PTMs conserved across different species as an evolutionary maintained response to specific biological stimuli?
Are viral pathogens tapping into lamin PTMs to control nuclear functions and gene expression?
ACKNOWLEDGEMENTS
We are grateful for funding from the National Institutes of Health NIGMS (GM114141) and the Edward Mallinckrodt Foundation to I.M.C., a National Science Foundation Graduate Research Fellowship (NSF-GRFP DGE-1656466) to L.A.M.N., and the NIH NIGMS (T32GM007388).
GLOSSARY
- Acetylation
the addition of an acetyl group to an amino acid side chain; neutralizes the positive charge of the amino acid
- Coiled-coil rod domain
alpha-helical domain of lamins that mediates lamin-lamin interactions and assembly into higher order filaments
- Farnesylation
the addition of an isoprenoid lipid to a cysteine side chain at a CaaX motif
- Head domain
N-terminal domain of lamins
- Homologous recombination
a method of double-strand break DNA repair that depends on extensive homology and is predominately active in S/G2 cell cycle phases
- Ig-like fold domain
C-terminal domain of lamins that mediates interactions with non-lamin proteins
- Interphase
phase of the cell cycle (including G1, S, and G2) during which a cell grows, replicates its genome, and prepares to divide
- Lamin
a type V intermediate filament that localizes at the inner nuclear membrane, maintains nuclear stability, and regulates nuclear processes
- Lamin associated domains (LADs)
heterochromatic DNA regions associated with the lamina at the nuclear periphery
- Laminopathies
genetic diseases caused by mutation in a lamin-encoding gene; examples are Emery-Dreifuss muscular dystrophy and Hutchinson-Gilford progeria syndrome
- Lysosomal degradation
degradation by the lysosome, a membrane-bound organelle that contains hydrolytic enzymes that can degrade proteins
- Methylation
the addition of a methyl group to an amino acid side chain
- Micronuclei
membrane-enclosed compartments containing fragmented genetic material or whole chromosomes
- Mitosis
the stage of the cell cycle in which one cell divides into two daughter cells with the same number of chromosomes as the parent cell
- Nonhomologous end joining
a double-strand break DNA repair pathway that functions throughout the cell cycle, depends on the direct ligation of broken DNA ends, and does not require extensive homology
- Nuclear-replicating virus
a type of virus that replicates its genome inside the host cell nucleus; examples are herpesviruses and adenoviruses
- O-GlcNAcylation
the addition of the sugar ß-O-linked N-acetylglucosamine to an amino acid side chain
- Phosphorylation
the addition of a phosphate group to an amino acid side chain
- Post-translational modification (PTM)
the enzymatic or non-enzymatic addition of a range of chemical functional groups or small proteins to amino acid side chains
- Prelamin A
the form of lamin A that is expressed prior to additional post-translational processing
- Progerin
a mutant form of lamin A in which 50 amino acids are deleted, causing Zmpste24 cleavage site removal and inducing permanent farnesylation; its expression causes progeria
- Proteasomal degradation
degradation by the proteasome, a multi-subunit protein complex that degrades proteins by breaking their peptide bonds
- SUMOylation
the addition of the small ubiquitin-like modifier (SUMO) protein to an amino acid side chain
- Ubiquitination
the addition of the small protein ubiquitin to an amino acid side chain; can target a protein for proteasomal degradation
- Viral capsid
a coat of virally-encoded proteins inside which the viral genome is packaged
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lin F and Worman HJ (1995) Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics 27, 230–236 [DOI] [PubMed] [Google Scholar]
- 2.Lin F and Worman HJ (1993) Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 268, 16321–16326 [PubMed] [Google Scholar]
- 3.Fisher DZ et al. (1986) cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. U. S. A. 83, 6450–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKeon FD et al. (1986) Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 319, 463–468 [DOI] [PubMed] [Google Scholar]
- 5.Broers JLV et al. (2006) Nuclear lamins: Laminopathies and their role in premature ageing. Physiol. Rev. 86, 967–1008 [DOI] [PubMed] [Google Scholar]
- 6.Dechat T et al. (2008) Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittmer T and Misteli T (2011) The lamin protein family. Genome Biol. 12, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke B and Stewart CL (2013) The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 14, 13–24 [DOI] [PubMed] [Google Scholar]
- 9.Heald R and McKeon F (1990) Mutation of phosphorylation sites in lamin A that prevent nuclear lamina dissembly in mitosis. Cell 61, 579–589 [DOI] [PubMed] [Google Scholar]
- 10.Pascual-Reguant L et al. (2018) Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 9, 3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gesson K et al. (2016) A-type Lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 26, 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaître C et al. (2014) Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Sandre-Giovannoli A et al. (2003) Lamin A truncation in Hutchinson-Gilford progeria. Science (80-. ). 300, 2055. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson M et al. (2003) Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423, 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padiath QS et al. (2006) Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet. 38, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 16.Hegele RA et al. (2006) Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am. J. Hum. Genet. 79, 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada T et al. (2014) Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irianto J et al. (2017) DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol. 27, 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ungricht R and Kutay U (2017) Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18, 229–245 [DOI] [PubMed] [Google Scholar]
- 20.Jia Y et al. (2019) Lamin B1 loss promotes lung cancer development and metastasis by epigenetic derepression of RET. J. Exp. Med. 216, 1377–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guelen L et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 22.Fu Y et al. (2015) MacroH2A1 associates with nuclear lamina and maintains chromatin architecture in mouse liver cells. Sci. Rep. 5, 17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux KJ et al. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickersgill H et al. (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 25.Tran JR et al. (2021) An APEX2 proximity ligation method for mapping interactions with the nuclear lamina. J. Cell Biol. 220, e202002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhary C et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (80-. ). 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 27.Weinert BT et al. (2018) Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundby A et al. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farnsworth CC et al. (1989) Human Lamin B contains a farnesylated cysteine residue. J. Biol. Chem. 264, 20422–20429 [PMC free article] [PubMed] [Google Scholar]
- 30.Rao RA et al. (2019) KMT 1 family methyltransferases regulate heterochromatin–nuclear periphery tethering via histone and non- histone protein methylation. EMBO Rep. 20, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen SC et al. (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 9, 1–15 [DOI] [PubMed] [Google Scholar]
- 32.Wang S et al. (2012) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates Akt signaling. PLoS One 7, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alfaro JF et al. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U. S. A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon D et al. (2018) OGT (O-GlcNAc Transferase) selectively modifies multiple residues unique to lamin A. Cells 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornbeck PV et al. (2015) PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen JV et al. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 37.Beausoleil SA et al. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinert BT et al. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 [DOI] [PubMed] [Google Scholar]
- 39.Lumpkin RJ et al. (2017) Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 8, 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner SA et al. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Povlsen LK et al. (2012) Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 14, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 42.Adam SA et al. (2013) Disruption of lamin B1 and lamin B2 processing and localization by farnesyltransferase inhibitors. Nucl. (United States) 4, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutz RJ et al. (1992) Nucleoplasmic localization of prelamin A: Implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl. Acad. Sci. U. S. A. 89, 3000–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon DN and Wilson KL (2013) Partners and post-translational modifications of nuclear lamins. Chromosoma 122, 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerace L and Blobel G (1980) The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell 19, 277–287 [DOI] [PubMed] [Google Scholar]
- 46.Ward GE and Kirschner MW (1990) Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61, 561–577 [DOI] [PubMed] [Google Scholar]
- 47.Peter M et al. (1990) In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61, 591–602 [DOI] [PubMed] [Google Scholar]
- 48.Goss VL et al. (1994) Identification of nuclear β(II) protein kinase C as a mitotic lamin kinase. J. Biol. Chem. 269, 19074–19080 [PubMed] [Google Scholar]
- 49.Eggert M et al. (1993) Identification of novel phosphorylation sites in murine A- type lamins. Eur. J. Biochem. 213, 659–671 [DOI] [PubMed] [Google Scholar]
- 50.Hocevar BA et al. (1993) Identification of Protein Kinase C (PKC) phosphorylation sites on human lamin B. J. Biol. Chem. 268, 7545–7552 [PubMed] [Google Scholar]
- 51.Thompson LJ et al. (1997) Identification of protein phosphatase 1 as a mitotic lamin phosphatase. J. Biol. Chem. 272, 29693–29697 [DOI] [PubMed] [Google Scholar]
- 52.Moriuchi T et al. (2016) Lamin A reassembly at the end of mitosis is regulated by its SUMO-interacting motif. Exp. Cell Res. 342, 83–94 [DOI] [PubMed] [Google Scholar]
- 53.Li Y et al. (2020) Nuclear envelope rupture and NET formation is driven by PKCα-mediated lamin B disassembly. EMBO Rep. 21, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godement M et al. (2021) Neutrophil Extracellular Traps in SARS-CoV2 related Pneumonia in ICU patients: The NETCOV2 study. Front. Med. 8, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kochin V et al. (2014) Interphase phosphorylation of lamin A. J. Cell Sci. 127, 2683–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leukel M and Jost E (1995) Two conserved serines in the nuclear localization signal flanking region are involved in the nuclear targeting of human lamin A. Eur J Cell Biol 68, 133–142 [PubMed] [Google Scholar]
- 57.Edens LJ et al. (2017) PKC-mediated phosphorylation of nuclear lamins at a single serine residue regulates interphase nuclear size in Xenopus and mammalian cells. Mol. Biol. Cell 28, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marmiroli S et al. (2009) A-type lamins and signaling: The PI 3-kinase/Akt pathway moves forward. J. Cell. Physiol. 220, 553–561 [DOI] [PubMed] [Google Scholar]
- 59.Cenni V et al. (2008) Lamin A Ser404 is a nuclear target of akt phosphorylation in C2C12 cells. J. Proteome Res. 7, 4727–4735 [DOI] [PubMed] [Google Scholar]
- 60.Buxboim A et al. (2014) Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 24, 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitsuhashi H et al. (2010) Specific phosphorylation of Ser458 of A-type lamins in LMNA-associated myopathy patients. J. Cell Sci. 123, 3893–3900 [DOI] [PubMed] [Google Scholar]
- 62.Krimm I et al. (2002) The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 10, 811–823 [DOI] [PubMed] [Google Scholar]
- 63.Karoutas A et al. (2019) The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol. 21, 1248–1260 [DOI] [PubMed] [Google Scholar]
- 64.Murray LA et al. (2018) Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat. Commun. 9, 4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YQ and Sarge KD (2008) Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J. Cell Biol. 182, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beck LA et al. (1988) Incorporation of a product of mevalonic acid metabolism into proteins of Chinese hamster ovary cell nuclei. J. Cell Biol. 107, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pendás AM et al. (2002) Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 31, 94–99 [DOI] [PubMed] [Google Scholar]
- 68.Wood KM et al. (2020) Defining substrate requirements for cleavage of farnesylated prelamin A by the integral membrane zinc metalloprotease ZMPSTE24. PLoS One 15, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jung HJ et al. (2013) Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl. Acad. Sci. U. S. A. 110, 1923–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon DN et al. (2013) Lamin A tail modification by SUMO1 is disrupted by familial partial lipodystrophy-causing mutations. Mol. Biol. Cell 24, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferraro A et al. (1989) Glycosylated forms of nuclear lamins. FEBS Lett. 257, 241–246 [DOI] [PubMed] [Google Scholar]
- 72.Miteva YV and Cristea IM (2014) A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol. Cell. Proteomics 13, 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milon BC et al. (2012) Role of histone deacetylases in gene regulation at nuclear lamina. PLoS One 7, e49692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santi S et al. (2020) PCAF involvement in lamin A/C-HDAC2 interplay during the early phase of muscle differentiation. Cells 9, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mattioli E et al. (2018) Altered modulation of lamin A/C-HDAC2 interaction and p21 expression during oxidative stress response in HGPS. Aging Cell 17, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ikegami K et al. (2020) Phosphorylated Lamin A/C in the nuclear interior binds active enhancers associated with abnormal transcription in progeria. Dev. Cell 52, 699–713.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boubriak II et al. (2017) Stress-induced release of Oct-1 from the nuclear envelope is mediated by JNK phosphorylation of lamin B1. PLoS One 12, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng X et al. (2018) Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell 71, 802–815.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murray-Nerger LA et al. (2021) Lamin B1 acetylation slows the G1 to S cell cycle transition through inhibition of DNA repair. Nucleic Acids Res 49, 2044–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mattioli E et al. (2019) Statins and histone deacetylase inhibitors affect Lamin A/C – histone deacetylase 2 interaction in human cells. Front. Cell Dev. Biol. 7, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cenni V et al. (2020) Lamin A involvement in ageing processes. Ageing Res. Rev. 62, 101073. [DOI] [PubMed] [Google Scholar]
- 82.Liu B et al. (2005) Genomic instability in laminopathy-based premature aging. Nat. Med. 11, 780–785 [DOI] [PubMed] [Google Scholar]
- 83.Lattanzi G et al. (2014) Lamins are rapamycin targets that impact human longevity: A study in centenarians. J. Cell Sci. 127, 147–157 [DOI] [PubMed] [Google Scholar]
- 84.Li Y et al. (2019) Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J. Exp. Clin. Cancer Res. 38, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dou Z et al. (2015) Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bertacchini J et al. (2013) The protein kinase Akt/PKB regulates both prelamin A degradation and Lmna gene expression. FASEB J. 27, 2145–2155 [DOI] [PubMed] [Google Scholar]
- 87.Shimizu T et al. (1998) Lamin B phosphorylation by protein kinase Cα and proteolysis during apoptosis in human leukemia HL60 cells. J. Biol. Chem. 273, 8669–8674 [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y et al. (2018) Nuclear Nestin deficiency drives tumor senescence via lamin A/C-dependent nuclear deformation. Nat. Commun. 9, 3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krishnamoorthy V et al. (2018) E3 ubiquitin ligase HECW2 targets PCNA and lamin B1. Biochim. Biophys. Acta - Mol. Cell Res 1865, 1088–1104 [DOI] [PubMed] [Google Scholar]
- 90.Khanna R et al. (2018) E3 ubiquitin ligase RNF123 targets lamin B1 and lamin-binding proteins. FEBS J. 285, 2243–2262 [DOI] [PubMed] [Google Scholar]
- 91.Borroni AP et al. (2018) Smurf2 regulates stability and the autophagic–lysosomal turnover of lamin A and its disease-associated form progerin. Aging Cell 17, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oh YS et al. (2010) Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell 9, 810–822 [DOI] [PubMed] [Google Scholar]
- 93.Mou F et al. (2007) US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81, 6459–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee C-P et al. (2008) Epstein-Barr Virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 82, 11913–11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meng Q et al. (2010) Simian Virus 40 T/t antigens and lamin A/C small interfering RNA rescue the phenotype of an Epstein-Barr Virus protein kinase (BGLF4) mutant. J. Virol. 84, 4524–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Milbradt J et al. (2010) Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285, 13979–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milbradt J et al. (2016) The prolyl isomerase Pin1 promotes the herpesvirus-induced phosphorylation-dependent disassembly of the nuclear lamina required for nucleocytoplasmic egress. PLoS Pathog. 12, 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cano-Monreal GL et al. (2009) Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology 392, 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X et al. (2017) Baculovirus infection induces disruption of the nuclear lamina. Sci. Rep. 7, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang T et al. (2019) Cellular p32 is a critical regulator of porcine circovirus type 2 nuclear egress. J. Virol. 93, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Speese SD et al. (2012) Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Biron KK (2006) Maribavir: A promising new antiherpes therapeutic agent. In New Concepts of Antiviral Therapy pp. 309–336 [Google Scholar]
- 103.Steingruber M and Marschall M (2020) The cytomegalovirus protein kinase pUL97: Host interactions, regulatory mechanisms and antiviral drug targeting. Microorganisms 8, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McSharry JJ et al. (2001) Inhibition of ganciclovir-susceptible and -resistant human cytomegalovirus clinical isolates by the benzimidazole L-riboside 1263W94. Clin. Diagn. Lab. Immunol. 8, 1279–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Höger TH et al. (1990) Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma 99, 379–390 [DOI] [PubMed] [Google Scholar]
- 106.Höger T et al. (1988) Amino acid sequence and molecular characterization of murine lamin B as deduced from cDNA clones. Eur J Cell Biol 47, 283–290 [PubMed] [Google Scholar]
- 107.Ben-Harush K et al. (2009) The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 386, 1392–1402 [DOI] [PubMed] [Google Scholar]
- 108.Turgay Y et al. (2017) The molecular architecture of lamins in somatic cells. Nature 543, 261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nmezi B et al. (2019) Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc. Natl. Acad. Sci. U. S. A. 116, 4307–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shimi T et al. (2008) The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shimi T et al. (2015) Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26, 4075–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sullivan T et al. (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen NY et al. (2018) Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. U. S. A. 115, 10100–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Funkhouser CM et al. (2013) Mechanical model of blebbing in nuclear lamin meshworks. Proc. Natl. Acad. Sci. U. S. A. 110, 3248–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coffinier C et al. (2011) Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 22, 4683–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Swift J et al. (2013) Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science (80-. ). 341, 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Denais CM et al. (2016) Nuclear envelope rupture and repair during cancer cell migration. Science (80-. ). 352, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dhe-Paganon S et al. (2002) Structure of the globular tail of nuclear lamin. J. Biol. Chem. 277, 17381–17384 [DOI] [PubMed] [Google Scholar]
- 119.Strelkov SV et al. (2004) Crystal structure of the human lamin A coil 2B dimer: Implications for the head-to-tail association of nuclear lamins. J. Mol. Biol. 343, 1067–1080 [DOI] [PubMed] [Google Scholar]
- 120.Ahn J et al. (2019) Structural basis for lamin assembly at the molecular level. Nat. Commun. 10, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lilina AV et al. (2020) Lateral A11 type tetramerization in lamins. J. Struct. Biol. 209, 107404. [DOI] [PubMed] [Google Scholar]
- 122.Ruan J et al. (2012) Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1. FEBS Lett. 586, 314–318 [DOI] [PubMed] [Google Scholar]
- 123.Pascual-Reguant L et al. (2018) Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 9, doi: 10.1038/s41467-018-05912-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Santi S et al. (2020) PCAF Involvement in Lamin A/C-HDAC2 Interplay during the Early Phase of Muscle Differentiation. Cells 9, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang K et al. (2016) Non-homologous end joining: Advances and frontiers. Acta Biochim. Biophys. Sin. (Shanghai). 48, 632–640 [DOI] [PubMed] [Google Scholar]
- 126.Haraguchi T et al. (2008) Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121, 2540–2554 [DOI] [PubMed] [Google Scholar]
- 127.Moir RD et al. (2000) Nuclear lamins A and B1: Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151, 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sharma K et al. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 129.Kettenbach AN et al. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Svinkina T et al. (2015) Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics 14, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim W et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hendriks IA et al. (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Song B et al. (2020) The DNA sensor cGAS is decorated by acetylation and phosphorylation modifications in the context of immune signaling. Mol. Cell. Proteomics 19, 1193–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karayel Ö et al. (2020) Accurate MS-based Rab10 phosphorylation stoichiometry determination as readout for LRRK2 activity in Parkinson’s Disease. Mol. Cell. Proteomics 19, 1546–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thomas SN et al. (2017) Targeted proteomic analyses of histone H4 acetylation changes associated with homologous-recombination-deficient high-grade serous ovarian carcinomas. J. Proteome Res. 16, 3704–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Morales Betanzos C et al. (2016) Dynamic phosphorylation of Apoptosis Signal Regulating Kinase 1 (ASK1) in response to oxidative and electrophilic stress. Chem. Res. Toxicol. 29, 2175–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mathias RA et al. (2014) Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang C et al. (2020) GPS 5.0: An update on the prediction of kinase-specific phosphorylation sites in proteins. Genomics Proteomics Bioinforma. 18, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Patrick R et al. (2017) PhosphoPICK-SNP: Quantifying the effect of amino acid variants on protein phosphorylation. Bioinformatics 33, 1773–1781 [DOI] [PubMed] [Google Scholar]