Figure 8.
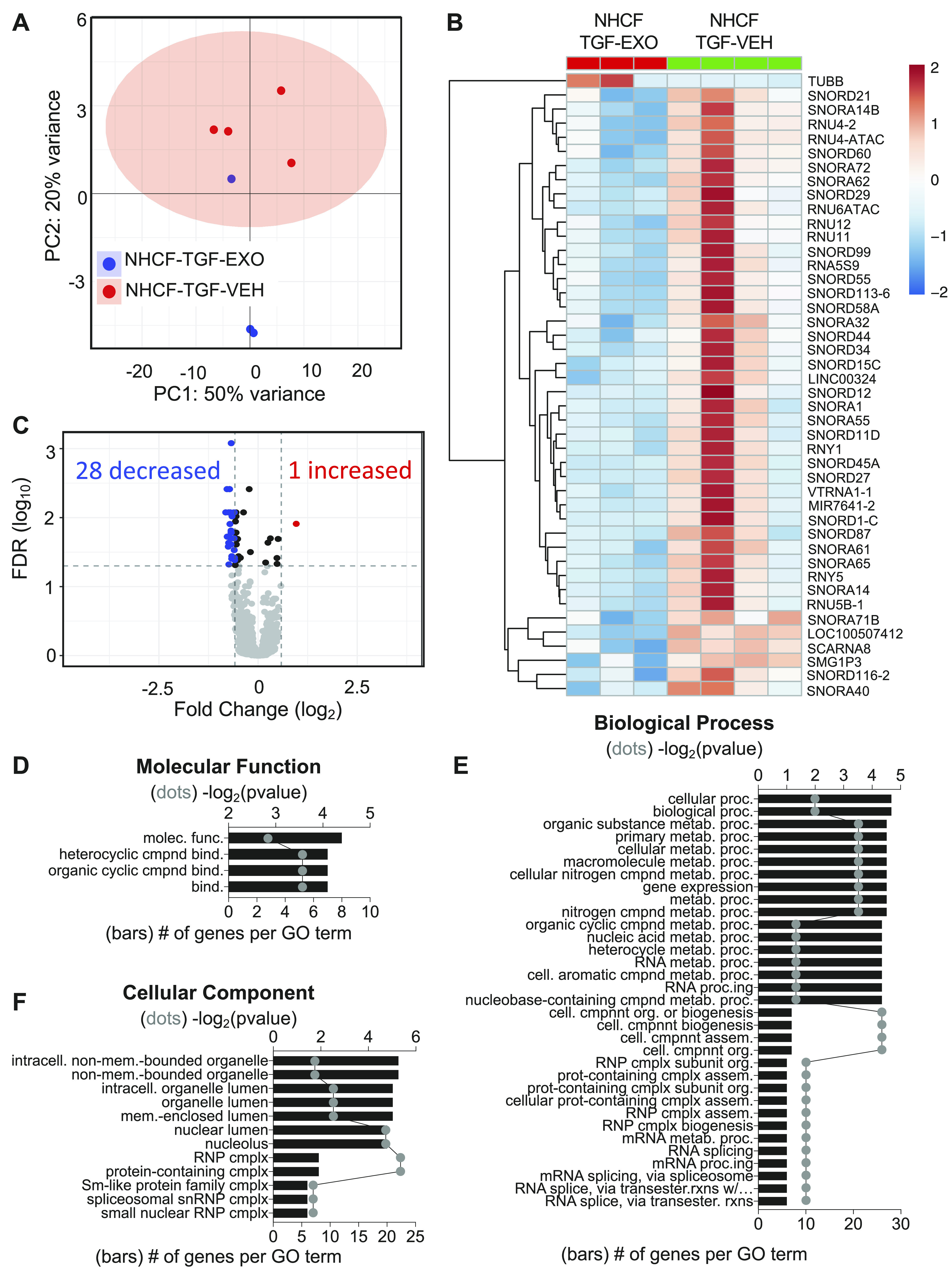
hCBSC-dEXO treatment of activated NHCFs leads to downregulation in miRNA and snoRNA related to ribosome stability and protein translation. Differential expression and gene ontology analysis of NHCFs activated with TGFβ to induce fibrosis and treated with either VEH or hCBSC exosomes. A: principal component analysis shows two distinct transcriptional profiles between NHCF-TGF-VEH and NHCF-TGFβ-EXO. B: heatmap of the 1 significantly increased transcript and 20 significantly deceased transcripts. C: volcano plot analysis showing transcripts with a FC ≥ |1.5| and FDR ≤ 0.05. Gene ontology analysis indicating the molecular function (D), biological process (E), and cellular components of genes (F) that are significantly differentially expressed (n = 4 for TGF-VEH; n = 3 for TGF-EXO). CBSCs, cortical bone stem cells; FC, fold change; FDR, false discovery rate; hCBSC-dEXO, human CBSC-derived exosome; NHCFs, normal human ventricular cardiac fibroblasts; snoRNA, small nucleolar RNA; TGFβ, transforming growth factor β; VEH, vehicle.
