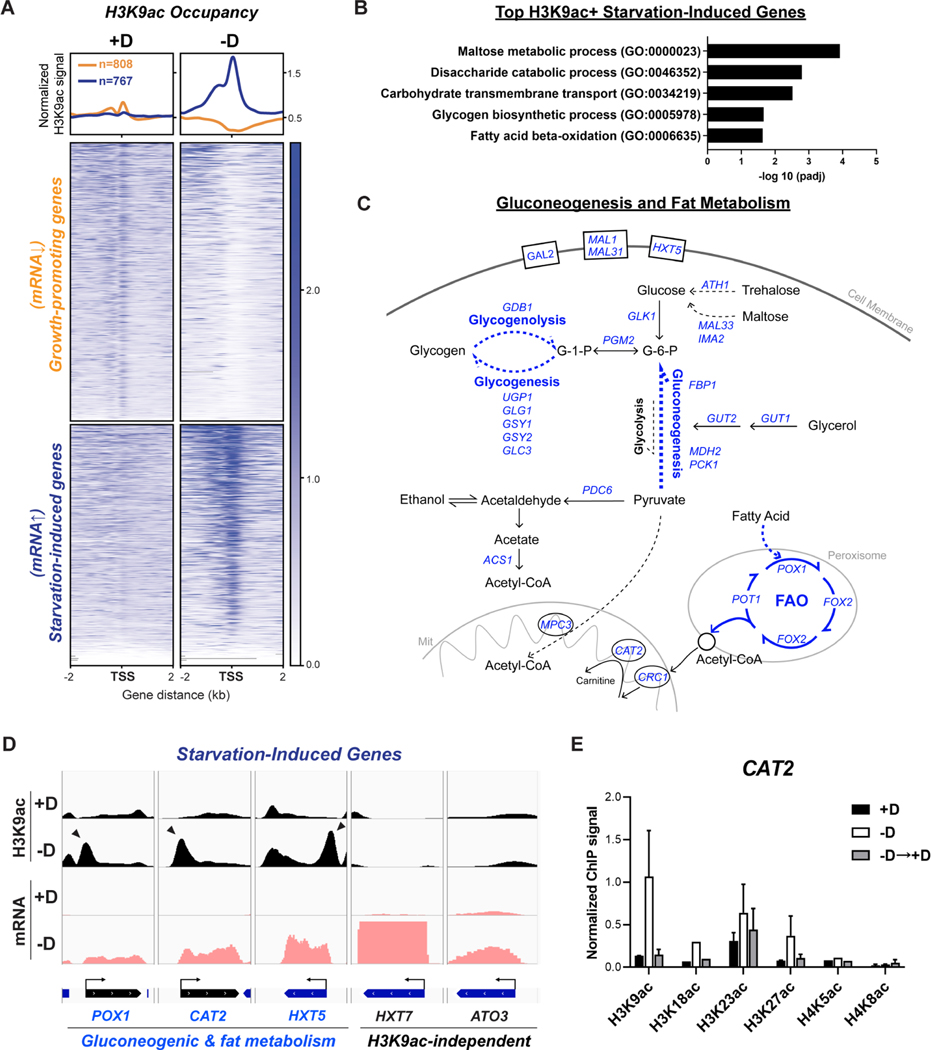
Figure 3. H3K9ac is enriched at the TSSs of genes required for gluconeogenesis and fat metabolism upon glucose starvation.
(A) ChIP-seq data displaying H3K9ac occupancy at two subsets of genes: growth-promoting genes (transcripts down-regulated upon starvation) and starvation-induced genes (transcripts up-regulated upon starvation). Metagene profile shows the average H3K9ac signal at either subset: growth-promoting genes in orange; starvation-induced genes in blue. Growth-promoting genes in heatmaps were ranked by signal intensity from glucose-replete condition (+D), while starvation-induced genes were ranked by signal intensity from glucose starvation (−D). Genes shown in rows are the same between samples.
(B) Gene ontology (GO) analysis of enriched biological processes for starvation-induced genes with most increased H3K9ac occupancy upon glucose starvation. See also Table S1.
(C) Consolidated enriched biological processes derived from GO analysis presenting major routes of carbon metabolic pathways in yeast under glucose starvation. Genes highlighted in blue were highly induced both in mRNA levels and H3K9ac signals upon glucose starvation.
(D) Genome browser view showing H3K9ac occupancy (in black) and mRNA levels (in pink) at starvation-induced genes under glucose-replete (+D) or glucose starvation (−D) conditions. The divergence of two subsets were differentiated based on the dependence on H3K9ac. H3K9ac-dependent genes involved in gluconeogenic and fat metabolism are shown in blue; H3K9ac-independent genes in black. Arrowheads indicate peaks of H3K9ac that reside at the TSS of genes of interest.
(E) ChIP-PCR analysis of histone acetylation occupancy at TSS of CAT2 under three conditions: glucose-replete (+D), glucose starvation for 30 min (−D), and glucose replenished for 30 min (−D→+D). All signals were normalized to H3. The data are represented as mean ± SD (n = 3).