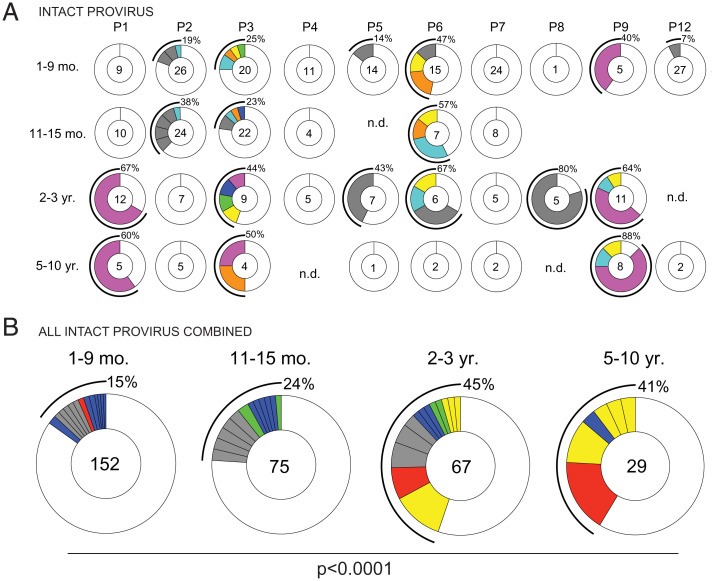
Fig. 5.
Clonal dynamics of the intact provirus reservoir over time. (A) Pie charts of intact proviruses detected in each individual participant over time. Participant name is denoted above the circles. Number inside each circle indicates the number of intact proviruses analyzed for each time point, specified to the Left of the circles. Color slices indicate persisting intact clones found at multiple time points, while gray slices are clonal expansions unique to the time point. Remaining white slices represent sequences isolated only once. Black outlines indicate the frequency of clonally expanded intact proviruses detected in each participant. n.d., not detected, no intact proviruses were detected from this sample. (B) Pie chart showing clonality of all intact proviruses detected in all individuals, combined, over time. Time point assays are denoted above the circles. Number inside each circle indicates the number of intact proviruses analyzed for each time point. Color slices indicate the following: Gray, clonally expanded intact proviruses unique to the time point assays; blue, persisting clones of intact provirus, first detected at 1 to 9 mo post-ART; green, persisting clones first detected at 11 to 15 mo post-ART; yellow, persisting clones first detected 2 to 3 y post-ART; red, persisting clones detected at all assayed time points; and white, intact provirus sequences isolated only once. Black line indicates the frequency of clonally expanded intact proviruses detected at each time point. A Fisher’s exact test with subsequent Bonferroni correction was used to compare clonality of intact proviruses between time points.