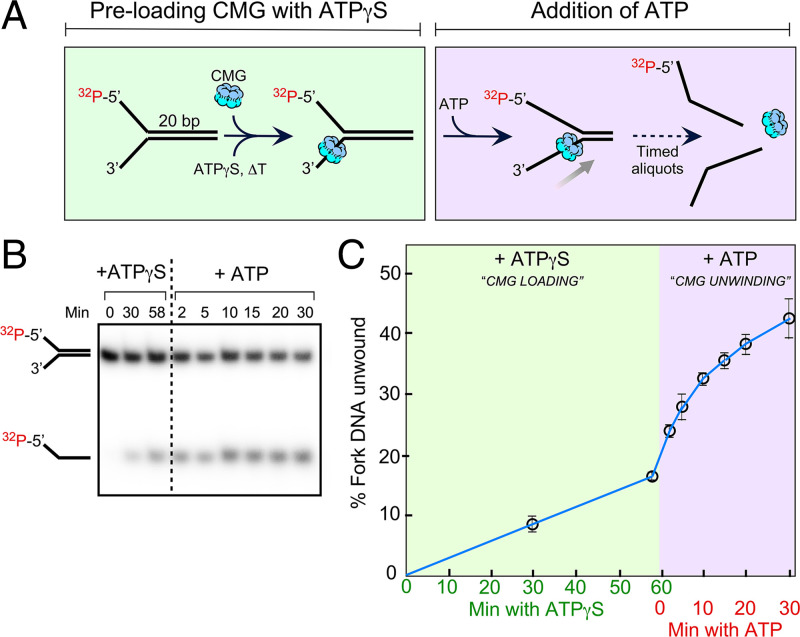
Fig. 1.
CMG can use ATPγS to unwind a DNA fork. (A) Unwinding of a 20-bp forked DNA by CMG. The scheme (Left) explains that the forked DNA is preincubated with CMG and 0.1 mM ATPγS for various times, and then 5 mM ATP is added to initiate unwinding (Right). (B) Native 10% polyacrylamide-gel electrophoresis (PAGE) of the helicase assays. Products formed during the preincubation time with ATPγS are shown in the Left three lanes, and reaction times with ATP are shown in the Right six lanes. (C) Quantitation of the gel data in B; error bars show the SEM.