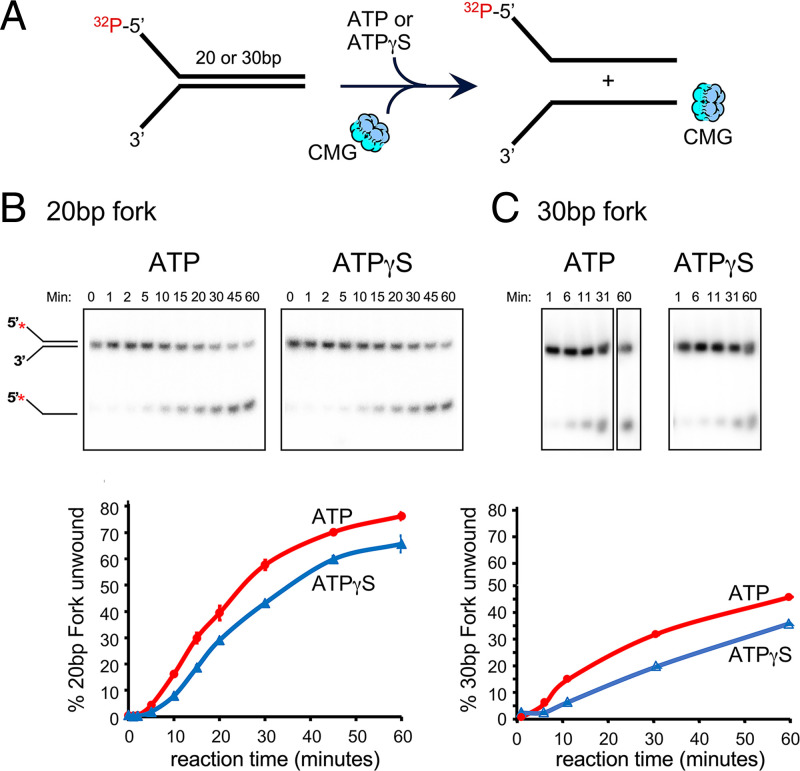
Fig. 2.
CMG unwinds 20- and 30-bp forked DNAs of different sequences at nearly the same rate using ATPγS or ATP. (A) Scheme of the assays. No preloading step was used. Instead, ATP or ATPγS was present and CMG was added directly to assays. (B) Native PAGE of CMG helicase assays using either ATP or ATPγS to unwind a 20-bp forked DNA assembled from Y20 leading and lagging oligos (SI Appendix, Table S1). (B, Bottom) Quantitation of the results; assays were performed in triplicate and error bars show the SEM. (C) Native PAGE of CMG helicase assays using either ATP or ATPγS, to unwind a 30-bp forked DNA having distinct sequences from the 20-bp fork, and assembled from N30 leading and lagging oligos. (C, Bottom) Quantitation of the gels is shown. Oligo sequences are in SI Appendix, Table S1.