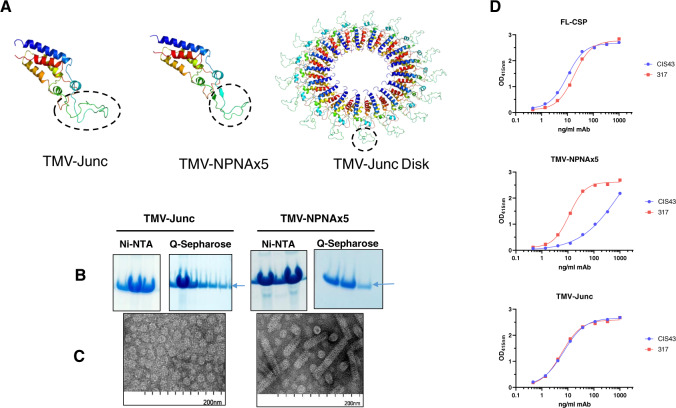
Fig. 1. Design and characterization of TMV-based constructs used in Study 1.
A Predicted monomer structures of circular permutants with inserted epitopes (circled). Left (TMV-Junc); middle (TMV-NPNAx5); and right (fully assembled TMV-Junc disk based on 3KML crystal structure). All structures were predicted by Rosetta Protein Prediction Server and images generated using PyMOL software. B Coomassie blue stained SDS-PAGE gels showing Ni-NTA and Q-Sepharose purification results for TMV-Junc and TMV-NPNAx5 proteins (arrow). C Electron micrographs of each purified vaccine preparation. D Representative ELISA titration curve of mAbs 317 (red) and CIS43 (blue) binding to FL-CSP, TMV-NPNAx5, and TMV-Junc (top to bottom).