Figure 3.
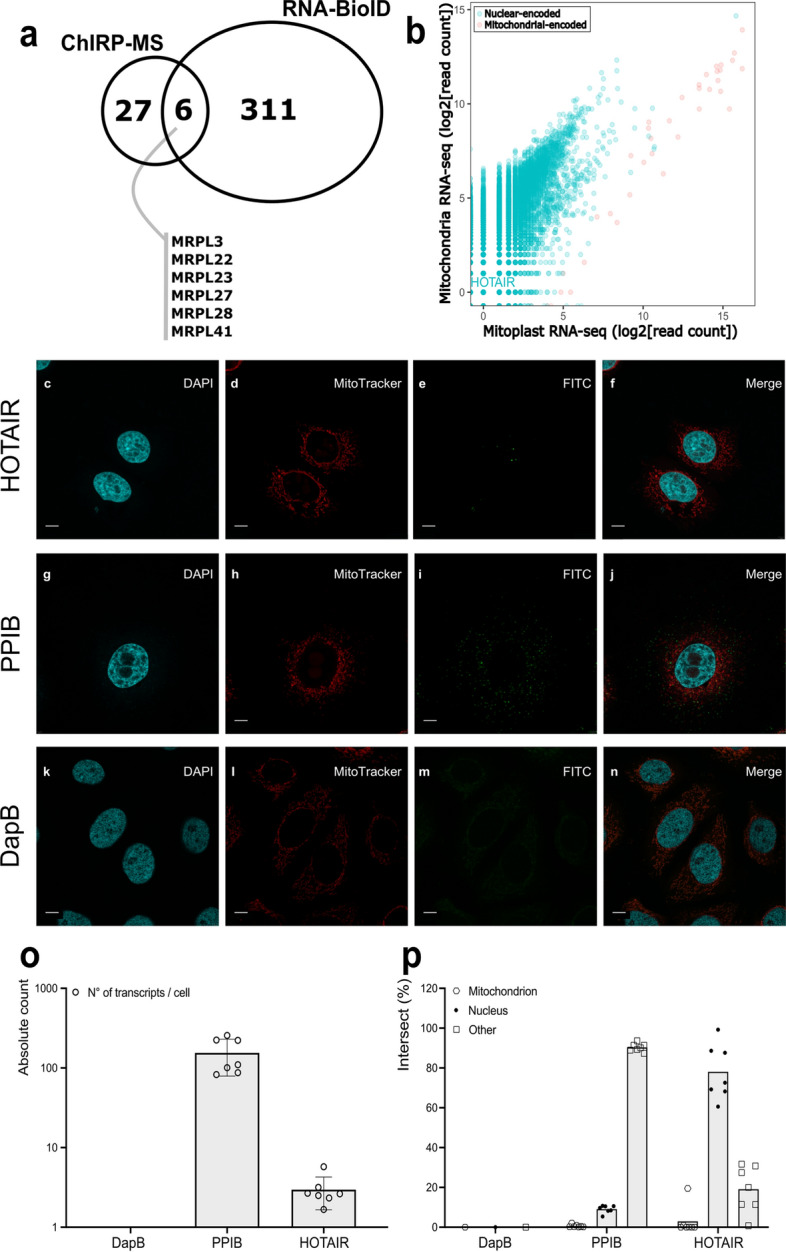
HOTAIR-MRPL interactions do not occur in mitochondria. (A) Overlap of ChIRP-MS and RNA-BioID identified proteins. Proteins identified in both methods are shown. (B) Reanalysis of RNA-sequencing data of Mercer et al. Nuclear-encoded genes are shown in green. Mitochondrial-encoded genes are shown in red. HOTAIR is highlighted. Colocalization of HOTAIR (C–F), PPIB mRNA positive control (G–J), and DapB mRNA negative control (K–N) with mitochondria in MCF7 determined by staining with RNAscope (FITC) and MitoTracker. (O) Determined transcript abundances per cell (n = 7) for each of the targets based on RNAscope puncti. (P) Intersect analysis (n = 7) showing percentage of the total transcript pool with mitochondria (hexagons) or nuclei (circles). Transcripts present in unstained organelle are shown as other (squares). The scale bar depicts 7.5 µm.
