Fig. 4.
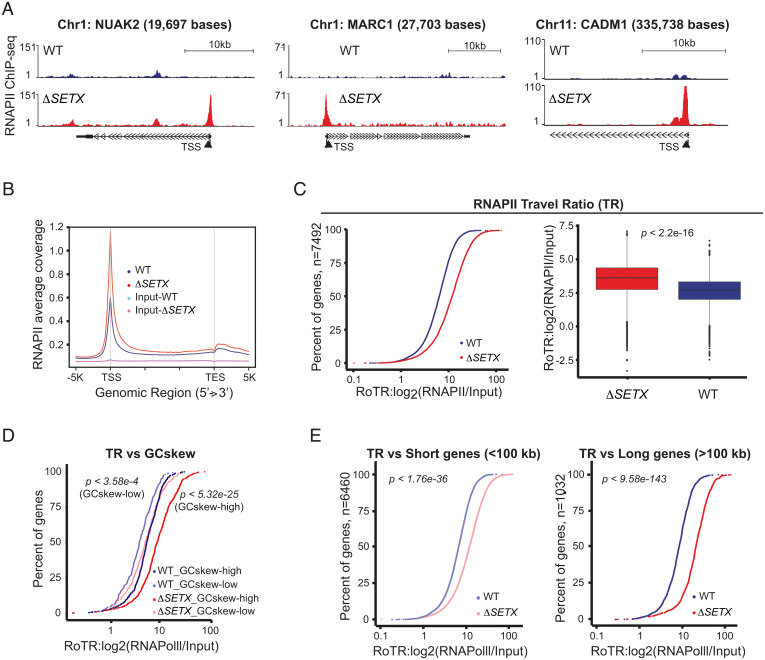
Loss of SETX causes transcription stress. (A) ChIP-seq analyses using RNAPII antibodies were carried out with cross-linked chromatin from HAP1 WT and ΔSETX cell lines. Representative genome browser shots of RNAPII binding over the NUAK2, MARC1, and CADM1 genes are shown. (Scale bars, 10 kb.) (B) Average RNAPII ChIP-seq coverage profiles for WT and ΔSETX are shown, along with input controls. TES, transcription end site. (C, Left) Cumulative curve of RNAPII ratio of traveling ratios (RoTR; x axis) for 7,492 genes in WT and ΔSETX. The y axis indicates percent of all genes. (Right) Box plots show the distribution of RoTR of 7,492 genes in WT and ΔSETX. P values were calculated using nonparametric Wilcoxon rank sum test. (D) Cumulative curve of RoTR in WT and ΔSETX with GCskew-high (n = 505) and GCskew-low (n = 215) groups that were called based on a 10% and 90% quantile from the GCskew frequency distribution of the entire genic intervals (n = 26,934). P values were calculated as in C. (E, Left) Cumulative curve of RoTR of 6,460 short genes (<100 kb) in WT and ΔSETX. (Right) Cumulative curve of RoTR of 1,032 long genes (>100 kb) in WT and ΔSETX. P values were calculated as in C.