Significance
Invading viral DNAs constitute a high risk for the infected cell and are high-profile targets of antiviral host factors. Nevertheless, little is known about the silencing machinery in the nucleus that acts to prevent transcription or retroviral integration of extrachromosomal DNA. We here identified CHAF1A and CHAF1B as two players that mediate silencing of unintegrated HIV-1 DNAs. Our findings provide evidence that these factors act independently of their canonical nucleosome assembly complex to induce silencing early in infection. The characterization of the intrinsic cellular defense mechanism against incoming DNA is relevant to transient gene delivery, as mediated by virus-based vectors in gene therapy.
Keywords: retroviral DNAs, HIV-1, transcriptional silencing, CHAF1A, histone chaperones
Abstract
Early events of the retroviral life cycle are the targets of many host restriction factors that have evolved to prevent establishment of infection. Incoming retroviral DNAs are transcriptionally silenced before integration in most cell types, and efficient viral gene expression occurs only after formation of the provirus. The molecular machinery for silencing unintegrated retroviral DNAs of HIV-1 remains poorly characterized. Here, we identified the histone chaperones CHAF1A and CHAF1B as essential factors for silencing of unintegrated HIV-1 DNAs. Using RNAi-mediated knockdown (KD) of multiple histone chaperones, we found that KD of CHAF1A or CHAF1B resulted in a pronounced increase in expression of incoming viral DNAs. The function of these two proteins in silencing was independent of their interaction partner RBBP4. Viral DNA levels accumulated to significantly higher levels in CHAF1A KD cells over controls, suggesting enhanced stabilization of actively transcribed DNAs. Chromatin immunoprecipitation assays revealed no major changes in histone loading onto viral DNAs in the absence of CHAF1A, but levels of the H3K9 trimethylation silencing mark were reduced. KD of the H3K9me3-binding protein HP1γ accelerated the expression of unintegrated HIV-1 DNAs. While CHAF1A was critical for silencing HIV-1 DNAs, it showed no role in silencing of unintegrated retroviral DNAs of mouse leukemia virus. Our study identifies CHAF1A and CHAF1B as factors involved specifically in silencing of HIV-1 DNAs early in infection. The results suggest that these factors act by noncanonical pathways, distinct from their histone loading activities, to mediate silencing of newly synthesized HIV-1 DNAs.
The early phase of the retrovirus life cycle is marked by the reverse transcription of viral RNA into linear double-stranded DNA, the entry of the DNA into the nucleus, and the integration of the viral DNA into the host genome to form the provirus (1). The late phase of the life cycle consists of the expression of viral genomic RNA, mRNAs, and viral protein precursors, and the production and release of new viral progeny followed by a final maturation step yielding fully infectious virions (2). The state of the newly synthesized retroviral DNA before integration remains poorly characterized. Unintegrated retroviral DNAs in the nucleus exist either as linear DNAs, which serve as the template for integration, or are circularized by the host to form circles containing either one copy of the long-terminal repeats (1-LTR circles) or two tandem copies (2-LTR circles) (3, 4). Viral DNAs that remain unintegrated do not replicate and gradually disappear over time, whereas integrated proviruses are retained and actively transcribed in most cell types. A remarkable feature of the unintegrated retroviral DNAs is that they are potently transcriptionally silenced (5–8). We have shown that the unintegrated DNAs of murine leukemia virus (MLV) are rapidly loaded with nucleosomes, and that these histones are marked by covalent modifications characteristic of silencing (9). We showed further that this silencing is mediated by the host protein NP220 together with the so-called HUSH (human silencing hub) complex (9, 10). In the case of HIV-1, we and others have shown that histones are also rapidly deposited onto unintegrated HIV-1 DNAs and marked with posttranslational modifications characteristic of silencing, and that transcriptional expression of these unintegrated forms of HIV-1 is efficiently suppressed (11, 12). There were significant differences in the host factors mediating the silencing: HIV-1 silencing was unaffected by HUSH knockout (10). The host cell machinery responsible for histone deposition onto histone-free extrachromosomal HIV-1 DNAs and for their silencing, however, is unknown.
In eukaryotic cells, nucleosomes are formed by the deposition of core histones H2A, H2B, H3, and H4 (13, 14). In addition to the canonical core histones, there exists a large variety of histone variants, such as histone variant H3.3, described to play a role in silencing endogenous retroviruses (15–17). Histones are deposited by specific histone chaperones. Histone chaperones are not only responsible for histone loading, but also for shaping the chromatin landscape by regulating histone availability and modifications (18, 19). Most histone chaperones exhibit specificities or preferences for the type of histones they deposit, and furthermore can be classified as mediating either replication-coupled or replication-independent histone deposition, such as during transcription or DNA repair (20). For example, the histone chaperones ATRX (ATRX chromatin remodeler), DAXX (death domain associated protein), DEK (DEK proto-oncogene), as well as the HIRA complex (formed by HIRA [histone regulator A], UBN1 [Ubinuclein1], and CABIN1 [calcineurin binding protein 1]) play important roles in deposition of the histone variant H3.3 during replication-independent histone loading (20). The histone chaperone NAP1L1 (nucleosome assembly protein 1 like 1) exhibits specificity for H2, while NASP (nuclear autoantigenic sperm protein) is reported to deposit H1 as well as canonical H3 (20). The histone chaperones CHAF1A (chromatin assembly factor 1 subunit A), CHAF1B (chromatin assembly factor 1 subunit B), and RBBP4 (retinoblastoma-binding protein 4) are components of the evolutionarily highly conserved CAF-1 (chromatin assembly factor 1) complex responsible for canonical H3 deposition (20, 21). In contrast to these loaders with restricted activities, the histone chaperone ASF1A (antisilencing function 1A histone chaperone) is involved in nucleosome assembly of both canonical H3 as well as histone variant H3.3 during replication-independent as well as replication-coupled histone deposition (20).
Here we have analyzed the impact of various histone chaperones on expression of unintegrated HIV-1 DNAs and their involvement in viral transcriptional silencing. We identified the histone chaperones CHAF1A and CHAF1B as previously unknown factors in silencing unintegrated HIV-1 DNAs. Our data suggest that these factors act by noncanonical pathways, distinct from their histone loading activities, to mediate silencing of newly synthesized HIV-1 DNAs early in infection.
Results
Histone Chaperones CHAF1A/B Mediate Silencing of Unintegrated HIV-1 DNAs.
We performed experiments to knockdown (KD) expression of several important histone chaperones (ATRX, DAXX, CABIN-1, ASF1A, NAP1L1, DEK, NASP, CHAF1A, CHAF1B) to explore their potential role in affecting viral expression of unintegrated HIV-1 DNAs. We used transient transfection of cells with small interfering RNAs (siRNAs) to induce efficient KD of endogenous levels of our candidate proteins. To achieve a specific readout of unintegrated DNAs of HIV-1, we utilized viral reporter constructs based on the pNL4-3.R−.E− HIV-1 strain, and compared a version containing a point mutation in the viral integrase to prevent integration (IN-D64A) with one containing the wild-type integrase (IN-wt). Both constructs encoded the fluorescence marker ZsGreen as a reporter of viral gene expression. KD cells were challenged with these reporter viruses, and ZsGreen expression was measured by flow cytometry 24 h after infection of either specific KD cells or control cells transfected with nontargeting siRNAs (NT-ctrl) (SI Appendix, Fig. S1A). We note that these reporters lack the viral Vpr gene, avoiding any of its potential effects on the host, such as cell cycle arrest (22–24). Of all the factors tested, two stood out for their ability to increase viral expression of unintegrated DNAs upon KD: CHAF1A and CHAF1B (SI Appendix, Fig. S1B). CHAF1A and CHAF1B are known to deposit newly synthesized and acetylated H3 and H4 histones in vitro as well as in vivo, and exhibit nucleosome assembly functions (25–27). The number of fluorescence-positive cells after infection with IN-D64A virus was dramatically increased up to sixfold with KD of either CHAF1A or CHAF1B alone, indicating that both factors were needed for efficient silencing (Fig. 1 A and B). The KD of CHAF1A gave the larger effect, KD of CHAF1B gave a smaller effect, and KD of both simultaneously showed an effect similar to or only slightly larger than that of CHAF1A KD alone. Importantly, the level of expression of integration-deficient virus in KD cells was equal to the level of expression of integration-proficient virus in NT-ctrl cells, illustrating the large extent of the relief in silencing (Fig. 1 B and C).
Fig. 1.
CHAF1A/B are required for silencing of unintegrated HIV-1 DNAs. (A) Cells were transfected with siRNAs targeting CHAF1A or CHAF1B, CHAF1A/B, or with a nontargeting siRNA control (NT-ctrl). Representative Western blots of KD and NT-ctrl cells probed with sera specific for the indicated proteins are shown. GAPDH was used as a housekeeping gene control. (B and C) KD or NT-ctrl cells were infected with integration-deficient (IN-D64A) or integration-proficient (IN-wt) ZsGreen reporter viruses, as indicated, using three different viral doses (undiluted virus, 1/3 and 1/10 dilution). Flow cytometry analyses were performed 24 h after infection. The percentages of ZsGreen+ cells are shown. Error bars indicate mean ± SD; n = 5 to 16 independent experiments. ns, not significant, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Mock represents uninfected control.
We performed experiments using several viral doses and found similar fold-increases in viral expression at all multiplicities of infection. ZsGreen expression of integration-proficient virus at 24 h after virus application, when cells contain both unintegrated and integrated viral DNAs, was also elevated in KD cells (Fig. 1C and SI Appendix, Fig. S1C), but with an increase of less than twofold. This smaller elevation could be attributed to the unintegrated DNA present in the IN-wt infections, though we cannot rule out some effect on expression from the integrated provirus as well. Once integration has occurred, expression becomes high with or without KD. The selectively enhanced viral gene expression of integration-deficient virus upon CHAF1A or CHAF1B KD was mainly manifest by an increase in the number of fluorescence-positive cells rather than an increase in the mean fluorescence intensity (MFI) of the ZsGreen+ cells (SI Appendix, Fig. S2). We conclude that CHAF1A and CHAF1B (hereafter CHAF1A/B) strongly suppress expression of unintegrated viral DNAs, and that upon CHAF1A/B depletion, viral expression is significantly activated.
Viral DNA Accumulates Significantly in the Absence of CHAF1A.
We next examined viral DNA levels at various time points after infection of KD and NT-ctrl cells with integrase-deficient and integration-proficient virus. The total levels of viral DNA were comparable in KD versus NT-ctrl cells at 6 h as well as 12 h after virus application, confirming that the virus preparations were at similar titers, that CHAF1A KD and control cells were equally susceptible to infection, and that there were no significant differences in the efficiency of cell entry or reverse transcription of viral RNA to DNA (Fig. 2A and SI Appendix, Fig. S3A). Interestingly, the total viral DNA levels became significantly increased at 24 h after infection in the absence of CHAF1A. The increase in total viral DNA in CHAF1A KD cells was only seen with integration-deficient virus, and not with integration-proficient virus (Fig. 2 C and D and SI Appendix, Fig. S3 C and D). We also examined the levels of 2-LTR circles by qPCR using specific primers. The levels of 2-LTR circles were comparable at 6 and 12 h after virus application in KD or NT-ctrl cells, indicating similar rates of nuclear entry (Fig. 2B and SI Appendix, Fig. S3B). Notably, the levels of 2-LTR circles are known to be elevated when integration is prevented (11, 28). As expected, we found increased levels of 2-LTR circles after infection with integration-deficient virus compared to integration-proficient virus (Fig. 2B and SI Appendix, Fig. S3B). But, in addition to this general increase, the levels of 2-LTR circles with the integration-deficient virus were substantially further elevated in CHAF1A KD cells at 24 h after infection, while the levels of 2-LTR circles of integration-proficient virus increased only modestly (Fig. 2 E and F and SI Appendix, Fig. S3 E and F).
Fig. 2.
The level of total viral DNAs, including 2-LTR circles, increases over time in the absence of CHAF1A. (A and B) Time-course analyses were performed with CHAF1A KD or NT-ctrl cells infected with integration-deficient (IN-D64A) reporter virus and qPCR analyses were used to determine levels of total viral DNA (A) or 2-LTR circles (B). Data are shown relative to GAPDH levels. (C–F) CHAF1A KD or NT-ctrl cells were infected with integration-deficient (HIV-1_IN-D64A) or integration-proficient (IN-wt) reporter viruses as indicated and DNA was isolated 24 h after infection. qPCR analyses were performed with ZsGreen-specific primers to measure total viral DNA (C and D) or with primers specific to the LTR junction region of 2-LTR circles (E and F). Data are depicted relative to GAPDH levels. ns, not significant, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001. Data are expressed as mean ± SD; n = 3 to 4 independent experiments. Mock represents uninfected control.
These data suggest that the higher levels of viral DNAs in CHAF1A KD cells at 24 h after infection likely results from increased stability of the DNAs, leading to their accumulation and longer persistence. Increased stability of the unintegrated DNAs in CHAF1A KD cells was correlated with their increased expression, and might be a consequence of the active transcription of the DNAs. Since unintegrated retroviral DNAs exist as linear DNA or 1-LTR or 2-LTR circles, we were curious if the proportion of 2-LTR circles compared to the total viral DNA shifts in the absence of CHAF1A. To address this question, we calculated the ratio of 2-LTR circles to total viral DNA. The ratio increased over time during the 6-, 12-, and 24-h time-course experiment, as expected, due to 2-LTR circle formation in the nucleus (SI Appendix, Fig. S3 G and H). The proportion of 2-LTR circles exhibited no major differences between KD and control cells, suggesting no changes in the 2-LTR circle formation preferences per se due to the CHAF1A KD. Rather, all forms of the viral DNA were increased by the KD. The increase in DNA may be a significant aspect of the increased reporter gene expression by simply providing more templates available for transcription.
CHAF1A/B Function Independently of the CAF-1 Complex.
The third component of the canonical CAF-1 complex, RBBP4, has been reported to inhibit transcription of integrated HIV-1 proviral DNA by directly binding to the HIV-1 LTR (29). In some settings, CHAF1A/B and RBBP4 are known to act independently from each other. A previous study identified CHAF1A/B, but not RBBP4, as responsible for epigenetic silencing of endogenous retroviruses, and RBBP4 is well-known to be part of various other complexes regulating histone metabolism as well as DNA repair (21, 30). To test for its involvement in silencing unintegrated HIV-1 DNAs, we performed KD of RBBP4 (SI Appendix, Fig. S4A) and applied either integration-deficient or integration-proficient virus to KD and NT-ctrl cells. Depletion of RBBP4 did not enhance viral expression but instead resulted in a modest inhibitory effect on expression from both viruses (SI Appendix, Fig. S4 B–D). The cause of the decrease in expression is unknown, but may reflect a general effect on transcription or any aspect of cell physiology. The effects were clearly distinctive from the increase seen with KD of CHAF1A/B. These data suggest that CHAF1A/B represses viral expression of unintegrated HIV-1 DNAs independently of the involvement of RBBP4.
Overall Histone Loading Is Not Affected by CHAF1A KD, But H3K9me3 Silencing Mark Levels Are Decreased.
CHAF1A/B are known to function in histone deposition onto DNA as well as to mediate chromatin rearrangements and changes in chromatin structure (21, 25). To test whether CHAF1A KD altered the histone deposition onto unintegrated HIV-1 DNAs, we performed chromatin immunoprecipitation (ChIP) experiments with antibodies specific to various histones and histone modifications and scored the proportion of bound viral DNA by qPCR. Our selections of histones and posttranslational histone modifications for examination were determined by the availability of suitable antibodies for ChIP assays. We used antibodies for H3 and H2B histones as classic members of core histone proteins, and used H1.2- and H1.4-specific antibodies to check for the presence of linker histones. The histone variant H3.3 was of interest because of its description as playing a role in silencing endogenous retroviruses (15–17). With regard to histone modifications, we chose H3K9me3 as a marker for inactive genes, and H3ac as a marker for active genes.
We found that the levels of unintegrated HIV-1 DNAs loaded with H3 and H2B core histones, H1 linker histones, and H3.3 histone variant were not significantly changed upon infection of CHAF1A KD cells compared to NT-ctrl cells (Fig. 3A; indicated as “ns”). The amount of viral DNA marked by histone H3 acetylation, a posttranslational mark characteristic for transcriptionally active DNA, was also similar in KD and NT-ctrl cells (Fig. 3A). The levels of the silencing mark H3K9me3 on DNA of integration-defective virus, however, were significantly reduced in KD cells compared to control cells (Fig. 3A). The levels of viral DNA marked by H3K9me3 were about twofold lower relative to the levels of DNAs bound to total H3 in KD cells, while these were roughly equal in control cells. Thus, CHAF1A is involved in promoting the introduction of the H3K9me3 silencing mark on viral chromatin. The histone profiles of the control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-globin genes confirmed that KD of the CHAF1A histone chaperones did not induce global changes to the cellular chromatin landscape (Fig. 3 B and C). We noted small increases in the levels of several histones at the globin gene upon KD but the significance of these changes is unclear.
Fig. 3.
CHAF1A KD does not alter histone loading onto unintegrated HIV-1 DNAs, but H3K9me3 formation is reduced. CHAF1A KD or NT-ctrl cells were infected with integration-deficient (IN-D64A) or integration-proficient (IN-wt) ZsGreen reporter viruses as indicated. ChIP assays with the sera specific for the indicated histones or histone modifications were performed 24 h after infection. Mouse IgG isotype control served as control for H2B-specific antibody; rabbit IgG was used as a control for all other histone-specific antibodies. DNA eluted from ChIP samples were analyzed by qPCR utilizing 2-LTR–specific primers (A), GAPDH-specific primers (B), or β-globin–specific primers (C). Data are shown as the percentage of input DNA and as mean ± SD; n = 2 to 6 independent experiments. ns, not significant, P ≥ 0.05; *P < 0.05.
A powerful tool in probing the chromatinization of DNA is the assay for transposase-accessible chromatin using sequencing (ATAC-seq), which tags open regions of DNA with short DNA oligonucleotides for PCR amplification, selecting for histone-free or less compacted DNA (31). We imagined that the method might reveal distinctions in the density or arrangement of nucleosomes on the viral DNA in CHAF1A/B KD cells as compared to control cells. To apply this method to monitor nucleosomal loading onto viral DNA, we infected either CHAF1A KD or control cells with integration-deficient or integration-proficient virus and at 24 h postinfection we performed transposase tagging, PCR amplification, and Illumina sequencing to generate more than 100 million reads per sample. We then mapped the reads to viral or host DNAs. The procedure generated an extraordinarily high frequency of DNAs derived from the unintegrated viral DNA, with ∼1% of all the sequencing reads mapping to the viral DNA (SI Appendix, Fig. S5A), roughly 100 times the true abundance of the viral DNA relative to cellular DNA as judged by qPCR. While the reads mapping to the human genome showed the size distribution typical of nucleosomal chromatin, with peaks of mono- and di-nucleosomes clearly visible, the reads mapping to the virus showed a continuous smooth spread of sizes (SI Appendix, Fig. S5B). There were no differences detected in the frequency and size distribution of the viral reads between the CHAF1A KD and control cells. Uninfected cells yielded essentially no reads mapping to the virus, as expected. There were also essentially no reads mapping to the plasmid vector used to generate the virus, confirming that there was no contaminating plasmid DNA in the virus preparations. Aligning the reads to selected host genes GAPDH and HB2A (hemoglobin subunit-α) as controls revealed the expected enrichment for open regions of DNA, while aligning the reads to the virus showed a uniform distribution along the entire length of the viral DNA (SI Appendix, Fig. S5 C–E). These results suggest that the assay is selectively scoring a highly accessible subset of the viral DNA that is not condensed into chromatin and constitutes a highly preferred substrate for the transposase. The yield from this open substrate overwhelms any signal from chromatinized viral DNA. Thus, while ATAC-seq has revealed the presence of this form of the viral DNA, it does not provide useful information about the histone-covered viral DNA.
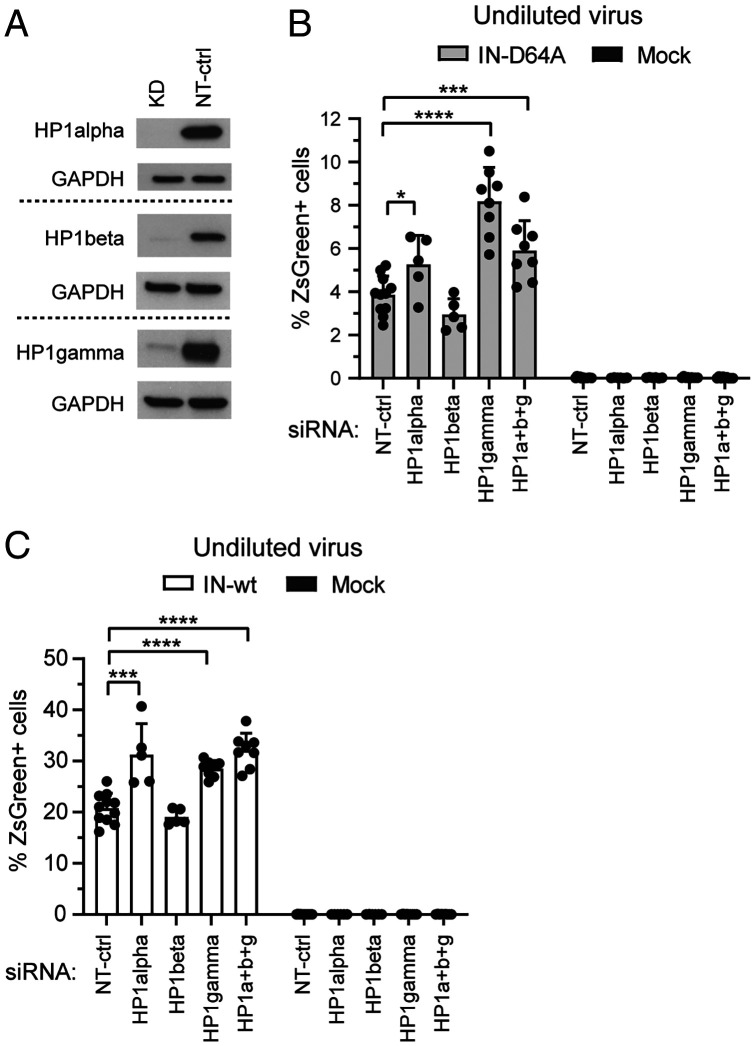
Viral Expression Is Increased in the Absence of the Heterochromatin Factor HP1γ.
Although CHAF1A/B first became prominent for its nucleosome assembly activity (25), the discovery of various interaction partners indicated broader functions, including roles in DNA replication, DNA repair and, of relevance here, maintenance of heterochromatin (21, 30). Of particular interest among its partners is the silencing factor HP1 (heterochromatin protein 1), a key player in heterochromatin formation (32–34). HP1 has a high binding affinity for H3K9me3 and genome-wide profiles revealed H3K9me3 enrichment at transcriptionally inactive endogenous retroviruses (35–37). HP1 has three different isoforms: HP1α (also known as CBX5), HP1β (CBX1), and HP1γ (CBX3). We performed single KDs of the different HP1 isoforms or a triple KD of all three isoforms (Fig. 4A) and measured viral expression 24 h after virus application. HP1γ KD and the triple HP1 KD consistently exhibited an approximately twofold increase in the number of fluorescence-positive cells upon infection with integration-deficient HIV-1 (Fig. 4B and SI Appendix, Fig. S6A). The MFIs were only slightly affected by the KD (SI Appendix, Fig. S6 C and E). Similar effects were seen with infection by integration-proficient virus in HP1γ KD and the HP1 triple KD cells, as well as HP1α KD cells (Fig. 4C and SI Appendix, Fig. S6 B, D, and F), but the effects were less pronounced than was seen with integration-deficient virus. Since both unintegrated and integrated forms of HIV-1 expression are present at 24 h after infection with integration-proficient virus, the effects on viral expression might well be largely or even exclusively derived from the unintegrated subset of HIV-1 DNAs.
Fig. 4.
KD of heterochromatin factor HP1γ increases expression of unintegrated HIV-1 DNAs. (A) Transient siRNA-based KD was performed for the three HP1 isoforms: HP1α, HP1β, HP1γ. Representative Western blots of HP1 proteins are shown for KD and NT-ctrl cells. GAPDH served as a housekeeping control. (B and C) Cells with single KDs of the three HP1 isoforms and triple KD of all three isoforms were infected with integration-deficient (IN-D64A) (B) or integration-proficient (IN-wt) ZsGreen reporter viruses (C). Flow cytometry analyses were conducted 24 h after infection. Data are expressed as mean ± SD; n = 5 to 11 independent experiments. ns, not significant, P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Mock represents uninfected control.
TRIM28 Has a Modest Effect on Expression of Unintegrated HIV-1.
TRIM28 (tripartite motif containing 28) is a major silencing cofactor, interacting with HP1, among other factors, and has been shown to induce transcriptional silencing of integrated DNA of MLV in embryonic stem cells, as well as of various endogenous retroviruses (35, 38, 39). Thus, it was of interest to analyze its potential role in silencing unintegrated HIV-1 DNAs. We conducted KD of TRIM28 (SI Appendix, Fig. S7A), applied integration-deficient or integration-proficient virus, and assessed ZsGreen expression 24 h after infection. In contrast to the strong effect of TRIM28 on silencing integrated MLV in embryonic stem cells, the impact on silencing unintegrated HIV-1 was minor. The percentage of fluorescence-positive cells was increased by up to twofold after infection with integration-deficient and slightly less with integration-proficient HIV-1, but the effect could only be seen using higher virus doses (SI Appendix, Fig. S7 B and C). As with KD of HP1 or CHAF1A/B, TRIM28 KD had no effect on the MFI of the ZsGreen+ cells (SI Appendix, Fig. S7D).
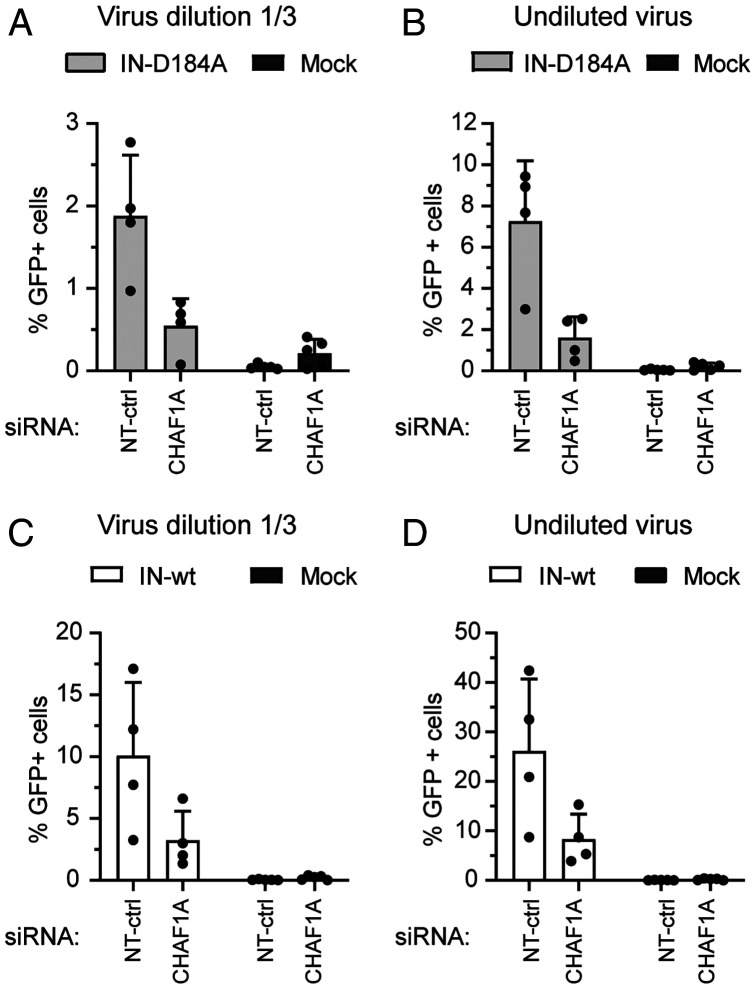
Expression of Unintegrated MLV DNA Is Not Altered by CHAF1A KD.
We next probed the specificity of CHAF1A function by examining the influence of CHAF1A KD on expression of the retrovirus MLV. CHAF1A KD cells were infected with either integration-deficient MLV (IN-D184A) or integration-proficient reporter virus (IN-wt) encoding green fluorescent protein (GFP). Interestingly, CHAF1A KD caused no increase in viral expression of either integration-deficient or integration-proficient MLV (Fig. 5 and SI Appendix, Fig. S8). Rather, there was a modest decrease in the number of fluorescence-expressing cells upon CHAF1A KD as compared to the control cells. These data give important evidence that CHAF1A/B does not mediate silencing of all incoming extrachromosomal or viral DNAs in general, but has a specific effect on HIV-1 DNAs. The data also indicate that CHAF1A/B are playing a complementary role to the HUSH complex, which was found to silence MLV but not HIV-1 DNAs (10). It remains unknown whether CHAF1A manifests selectivity for HIV-1 by specific interactions with HIV-1 DNA itself or through interaction with sequence-specific DNA-binding factors such as zinc-finger proteins. The selectivity could alternatively be attributable to the substantial differences in nuclear entry between HIV-1 and MLV. MLV depends on the disassembly of the nuclear membrane to enter the nucleus, whereas HIV-1 engages the nuclear pore complex to mediate its active entry, independent of mitosis (40–44). These two very different nuclear entry routes might well determine the differential access to host cofactors and regulatory machinery acting on viral DNA before integration.
Fig. 5.
CHAF1A KD has no positive effect on expression of unintegrated MLV DNAs. (A–D) CHAF1A KD or NT-ctrl cells were infected with integration-deficient (IN-D184A) or integration-proficient (IN-wt) GFP reporter viruses as indicated. Data from two different viral doses are shown (undiluted virus and 1/3 dilution). Flow cytometry analyses were conducted 24 h after infection and the percentages of GFP+ cells are depicted. Data are expressed as mean ± SD; n = 4 to 5 independent experiments. Mock was used as an uninfected control.
Discussion
Here, we identify CHAF1A/B as key factors required for silencing of unintegrated HIV-1 DNAs. CHAF1A/B KD results in a dramatic increase in expression of integration-deficient HIV-1 DNA (up to sixfold) and only minor increase in integration-proficient HIV-1 DNA (less than twofold). This relief of silencing is the highest fold-increase in expression from unintegrated HIV-1 DNAs known to date [as compared with KD of SMC 5/6 (45)]. It is notable that the activation of expression upon KD is mainly manifest as an increase in the number of cells expressing the fluorescence reporter, and not as much an increase in the MFI of those cells that have turned on reporter expression. The result suggests that CHAF1A/B are controlling a “decision” of the cell to express the virus or not, and are not affecting the magnitude of expression once it is turned on. We note that when integration is allowed, then there is an increase in the MFI, with or without the KD. CHAF1A/B KD may have multiple modes of action. The levels of the silencing mark H3K9me3 bound to unintegrated HIV-1 DNA are reduced in CHAF1A KD cells compared to the control, but this effect alone probably does not account entirely for the strong relief of silencing viral expression. It is likely that other aspects of the chromatin structures formed on the viral DNA are subtly changed by the KD. Nucleosomes may be reorganized or mobilized, allowing access to transcription machinery.
An unexpected observation was the finding that the amount of all viral DNAs, including the 2-LTR circles, significantly increases over time after virus infection of CHAF1A/B KD cells. This might be in some way attributable to the DNAs undergoing active transcription, and if true, this would suggest that both linear and circular forms are serving as templates for transcription. It could nevertheless be that one or the other form is a preferred template. The increase in stability and overall viral DNA levels might well contribute to the enhanced viral expression by providing additional viral templates for transcription. The fold-increase in expression is significantly greater than the increase in DNA levels, however, so changes in the conformation or state of the DNA must be involved. We also identified HP1γ, and to some extent TRIM28, as modestly contributing to the silencing of unintegrated HIV-1 DNAs. HP1γ might act to promote the spreading of the H3K9me3 mark along the viral DNA after an initial introduction of the mark by SETDB1 or other methyltransferases. TRIM28, however, only played a minor role in silencing unintegrated HIV-1 DNAs, in contrast to its major role in transcriptional silencing of MLV proviruses in embryonic stem cells through targeting the primer binding site (38). It is unknown what factor might place an initial H3K9me3 mark on the unintegrated DNA and whether this is a sequence-specific event.
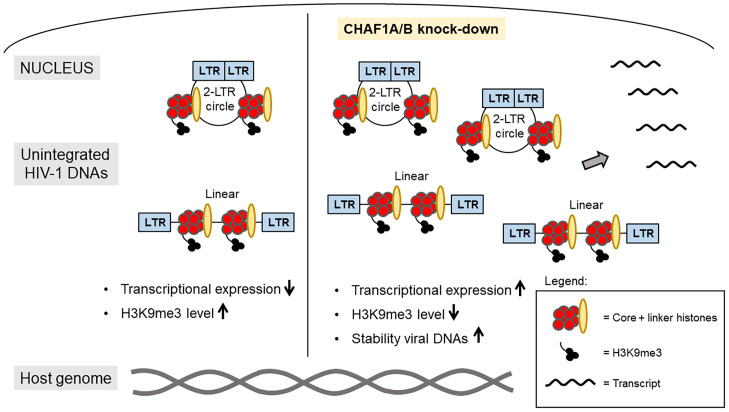
The results here demonstrate that the CHAF1A/B chromatin assembly factors play a major role in mediating the initial silencing of incoming HIV-1 DNAs before integration into the host genome (Fig. 6). KD of either factor results in substantial relief of the normal silencing and abnormally high gene expression. The CHAF1A/B silencing activity does not require their cofactor RBBP4, and thus is apparently not functioning in the context of the canonical CAF-1 complex. We found that these factors are responsible for silencing unintegrated DNAs of HIV-1, but not those of another retrovirus, MLV. They thus have the complementary activity to the HUSH complex, which is required for silencing MLV DNA but not HIV-1 (10). We are led to the realization that the machinery utilized by cells to suppress expression of a particular foreign DNA is not universal but varies with its origin or route of entry, or trafficking pathway, and presumably reflect different sequence elements recognized on the DNA. The machinery may also vary among cell types.
Fig. 6.
Schematic overview of the relief of silencing of unintegrated HIV-1 DNAs after CHAF1A KD. Unintegrated HIV-1 DNAs exhibit very poor transcriptional expression in wild-type cells (Left). ChIP analyses reveal deposition of core as well as linker histones and high level of the silencing mark H3K9 trimethylation on unintegrated HIV-1 DNAs. Upon KD of CHAF1A/B, viral expression increases significantly (Right). ChIP analyses indicate no major changes of histone loading, but decreased level of H3K9me3. Time-course experiments show an increased accumulation of viral DNAs 24 h after infection of KD cells.
It has been reported that the viral Vpr protein can relieve silencing of unintegrated HIV-1 DNAs (8, 45, 46), and since our reporter genomes lacked Vpr, it was of interest to test the effects of including Vpr. We performed infections with HIV-1 genomes expressing a luciferase reporter, with or without Vpr, and assayed luciferase expression in the presence or absence of the integrase inhibitor raltegravir. We found the unintegrated DNAs to be profoundly silenced even when Vpr was present (SI Appendix, Fig. S9), indicating that Vpr did not phenocopy the KD of CHAF1A/B. There was thus no indication in our assays that Vpr was inhibiting or mediating degradation of CHAF1A/B. We note that some reported effects involved expression of Vpr provided in trans rather than as delivered by the infecting virions.
Various DNA viruses are known to be silenced early in infection, and many viruses even encode factors that counteract the transcriptional silencing of their DNAs, such as the ICP0 protein of herpes simplex virus type 1 (47–50) and the HBx protein of hepatitis B virus (51). The HTLV-1 Tax protein, by recruiting NF-κB, can also overcome the silencing of unintegrated HIV-1 DNA (52, 53). This arms race between silencing factors and antisilencing factors reflects the importance of the early expression of viral genes and the advantages this provides for virus transmission and the course of infection. We note that there may also be settings in which the silencing is beneficial to the virus: it may increase the frequency with which the virus establishes latent proviruses, ultimately promoting the long-term survival of the virus in an infected host.
We expect that more silencing factors are yet to be identified. The chromatin remodelers SMC5/6, and their localization factor SLF2, have recently been shown to play a role in silencing of unintegrated HIV-1 DNA (45). We do not yet know if CHAF1A/B and SLF2, or other factors, work together or independently in silencing. Our data suggest that the CHAF1 proteins do not act at the time of loading of histones onto the incoming HIV-1 DNA per se, but rather at a later step in gene expression, at stages coupled to histone modifications, or chromatin rearrangements or condensation associated with silencing. The enhanced transcription upon KD is also associated with an increased accumulation of the unintegrated DNA, which may enhance expression in a positive feedback loop.
A full understanding of the silencing machinery of early HIV-1 events may suggest new methods of control of viral transmission through enhancing the efficiency of viral suppression and may offer implications for other steps of the HIV-1 life cycle. The CAF-1 complex became of particular interest for the HIV-1 field very recently, since it has been published that CAF-1 is enriched on HIV-1 LTRs and shown to promote maintenance of HIV-1 latency, a major hurdle to cure AIDS (34). Enhancing its activities might promote long-term silencing, while inhibition might promote reactivation and allow for ultimate virus clearance.
Finally, we note that our findings also have the potential to improve many gene-delivery applications, especially the use of HIV-1–based nonintegrating retroviral vectors in gene therapy, through inhibiting the silencing of the incoming vector DNAs.
Materials and Methods
Retrovirus Reporter.
HIV-1–based reporter virus encoded on plasmid pNL4-3.ZsGreen.R−.E− was used throughout the study (11). The integration-deficient version is here designated as IN-D64A, and the wild-type integration-proficient version as IN-wt. The corresponding Moloney MLV-based reporter virus on plasmid pNCA-GFP with an integrase mutation is here designated as IN-D184A, and the wild-type integration-proficient version as IN-wt (11). Viral particles were pseudotyped with the vesicular stomatitis virus (VSV) glycoprotein, encoded by pMD2.G (a gift from Didier Trono, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; Addgene plasmid, 12259).
Cells and Cultivation.
Human HeLa cells (American Type Culture Collection, CCL-2) and human Lenti-X 293T cells (Clontech, 632180) were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. KD cells were cultured in DMEM and fetal bovine serum in the absence of antibiotics.
Retrovirus Packaging and Infection.
Virus production in 293T cells was induced by transfection using the calcium phosphate precipitation method and as previously described (11). Virus supernatants were concentrated (100×) by ultracentrifugation (2 h, 25,000 rpm, 4 °C). Integrase mutant and integrase wild-type virus reporters were produced simultaneously by transfections and concentrations performed in parallel. Virus doses were indicated as undiluted, 1/3 and 1/10 diluted. Infections of control cells with undiluted virus preparations of the IN-wt virus resulted in ∼20 to 30% fluorescence-positive cells, which corresponds to a multiplicity of infection of ∼1. Equal amounts of IN-D64A or IN-wt were applied to cells for comparative studies. Next, 5 × 104 cells were seeded 1 d before infection in a 12-well format. Cells were incubated with virus particles for 5 h.
siRNA Transfection and Western Blot.
The siRNAs used throughout this study were purchased from Dharmacon with ON-TARGET plus SMART pool design (ATRX #L-006524-00; DAXX #L-004420-00; CABIN1 #L-012454-01; ASF1A #L-020222-02; NAP1L1 #L-017274-01; DEK #L-003881-00; NASP #L-011740-01; CHAF1A #L-019938-00; CHAF1B #L-019937-00; RBBP4 #L-012137-00; TRIM28 #L-005046-00; HP1α #L-004296-00; HP1β #L-009716-00; HP1γ #L-010033-00; nontargeting pool #D-001810-10). HeLa cells were seeded (4 × 105 cells per 10-cm dish) 1 d before transfection. siRNA transfection was performed with 0.3 nmol siRNA per 10-cm dish and mixed with Lipofectamine RNAiMAX (Life Technologies, 137785000), according to the manufacturer’s protocol. Transfection was performed on 2 consecutive days. Cells were seeded for infection at the end of the second siRNA transfection, and virus particles applied to KD cells the next day. KD cells were harvested for ChIP, flow cytometry or Western blot analyses ∼24 h after infection. Western blots were performed as previously described (11).
The antibodies and concentrations used were as follows: anti-ATRX ([H-300]; #sc-15408; Santa Cruz Biotechnology) 1:200; anti-DAXX ([M-112]; #sc-7152; Santa Cruz Biotechnology) 1:5,000; anti-CABIN1 ([D2B9F]; #12660; Cell Signaling Technology) 1:1,000; anti-ASF1A ([C6E10]; #2990; Cell Signaling Technology) 1:1,000; anti-NAP1L1 ([EPR12619]; #ab174836; Abcam) 1:1,000; anti-DEK ([E1L3V]; #13962; Cell Signaling) 1:1,000; anti-NASP ([3A301-011A]; Bethyl Laboratories) 1:1,000; anti-CHAF1A ([EPR5576(2)]; #ab126625; Abcam) 1:1,000; anti-CHAF1B ([EPR6105]; #ab109442; Abcam) 1:10,000; anti-RBBP4 (#A301-206A; Bethyl Laboratories) 1:1,000; anti-TRIM28 ([BL-248–2G6]; #A700-014; Bethyl Laboratories) 1:1,000; anti-HP1α (#2616S; Cell Signaling Technology) 1:1,000; anti-HP1β (#2613S; Cell Signaling Technology) 1:1,000; anti-HP1γ (#2619S; Cell Signaling Technology) 1:1,000; anti-GAPDH ([6C5]; #CB1001; Millipore-Sigma) 1:10,000; Amersham ECL rabbit IgG HRP-linked whole Ab (#NA934; Cytiva) 1:10,000; and Amersham ECL mouse IgG HRP-linked whole Ab (#NA931; Cytiva) 1:10,000.
Flow Cytometry Analysis.
Samples were measured with BD LSR II flow cytometer (BD Biosciences) and data analyzed with FlowJo software (BD Biosciences). Gating strategy included gating of FSC-H/SSC-H population for viable cells followed by FSC-H/FITC-H for ZsGreen+ cells. MFI was determined for the ZsGreen+ population.
Viral DNA Detection.
DNA was purified with QIAamp DNA blood mini kit (Qiagen) according to the manufacturer’s instructions. DNA concentrations were adjusted to 50 ng/µL and ∼50–100 ng DNA per sample was used for qPCR analyses based on SYBR Green (Roche, 4913850001). The following primers were used: 2-LTR circle detection (for 5′-AACTAGGGAACCCACTGCTTAAG-3′, rev 5′-TCCACAGATCAAGGATATCTTGTC-3′) and for ZsGreen as a readout for total viral DNA (for 5′-CCCCGTGATGAAGAAGATGA-3′, rev 5′-GTCAGCTTGTGCTGGATGAA-3′). qPCRs were performed with 7500 Fast Real-Time PCR System (Applied Biosystems) (PCR program: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C). A melting curve for quality control was generated for every run following the manufacturer’s advice. The Ct-values were normalized to endogenous GAPDH levels (for 5′-CAATTCCCCATCTCAGTCGT-3″, rev 5′-TAGTAGCCGGGCCCTACTTT-3′) using the 2ΔCt method.
ChIP Assay.
Cells were seeded (1.5 to 2 × 106 per 10-cm dish) one day before infection. Virus supernatants (15 µL) were pretreated with 5 U/mL DNase I (Promega, M6101) for 1 h at 37 °C. Medium was supplemented with 8 µg/mL Polybrene (Millipore-Sigma, TR-1003-G). Virus and polybrene were removed after 5 h. Cells were washed twice and harvested for ChIP ∼24 h after infection. ChIP protocol was performed as previously described (11). Next, ChIP-grade antibodies were used with a concentration of 5 µg per 50 µg sonicated chromatin: anti-H1.2 ([EPR12690]; #ab181973; Abcam); anti-H1.4 ([D4J5Q]; #41328S; Cell Signaling Technology); anti-H2B (#ab52484; Abcam); anti-H3 (#ab1791; Abcam); anti-H3.3 ([EPR17899]; #ab176840; Abcam); anti-H3K9me3 (#ab8898; Abcam); anti-H3 acetyl K9+K14+K18+K23+K27 (#ab47915; Abcam); rabbit IgG isotype control (#02–6102; Invitrogen); and mouse IgG1 isotype control ([G3A1]; #5415S; Cell Signaling Technology). After DNA purification, 5 µL per sample were used for qPCR analysis. PCR protocol and primers were used as described above in “Viral DNA Detection.” β-Globin–specific primers served as a heterochromatin control (for 5′-CAGAGCCATCTATTGCTTAC-3′, rev: 5′-GCCTCACCACCAACTTCATC-3′). Data were calculated relative to the respective input DNA with the 2ΔCt method and shown as percentage relative to input DNA in the bar graphs.
ATAC-seq.
CHAF1A KD cells were harvested 24 h after infection. Lysis, Tn5 transposition reactions and DNA purification were performed as previously published by Buenrostro et al. (31). The Tagment DNA Enzyme and Buffer kit from Illumina was used to mediate the DNA targeting. Qubit and Bioanalyzer measurements of the DNA were done for every sample before sequencing as quality controls. Paired-end sequencing was performed on the Illumina NextSEq. 550 sequencer. ATAC-seq reads were mapped to the HIV-1 genome and human genome assembly hg38 using HISAT2 (v2.1.0, parameter: -X 2000). Potential PCR duplicates were removed by the function “MarkDuplicates” (parameter: REMOVE_DUPLICATES = true) of Picard (v2.23.1). Genomic enrichment of ATAC-seq signals were visualized using the Integrative Genomics Viewer. The size distribution of fragments was measured by the function “CollectInsertSizeMetrics” of Picard (v2.23.1).
Construction of Luc Reporters and Luciferase Assay.
The wild-type Vpr sequence was excised from pNL4.3 (ARP-3418, NIH AIDS Reagent Program) by digestion with PflMI and SalI restriction enzymes and ligated into pNL4-3.Luc.E−R− (ARP-114; NIH AIDS Reagent Program), digested in the same manner. Virus was prepared by transfection of 293T cells with these DNAs with a VSV-G expression construct. Before infection, the viral supernatants were checked for the presence and absence of Vpr by Western blot. Anti-Vpr polyclonal antibody (Proteintech, #51143-1-AP) and anti-HIV1 p24 antibody [39/5.4A] (Abcam, #ab9071) were used for staining. HeLa cells were treated with 10 µM Raltegravir (Santa Cruz Biotechnology, sc-364600) or the equivalent amount of dimethyl sulfoxide and infected with reporter viruses encoding Vpr or without Vpr. Luciferase activity was assessed 24 h after infection with the Luciferase Assay System (Promega, #E1501) according to the manufacturer’s instructions and measured on an Omega microplate reader (BMG Labtech).
Statistical Analysis.
Data were shown as mean ± SD. Statistical method and sample size were used as indicated in the figure legends. The unpaired t test for comparison of only two groups, and additionally Welch’s correction, was chosen, if variances were significantly different between the two groups. Significance was indicated as follows: not significant (ns): P ≥ 0.05; significant: *P = 0.01 to 0.05; very significant: **P = 0.001 to 0.01; extremely significant: ***P = 0.0001 to 0.001; and extremely significant: ****P < 0.0001.
Supplementary Material
Acknowledgments
S.P.G. is an investigator of HHMI. This work was also supported in part by NIH Grants R01 CA030488 and R35GM138181. F.K.G. was supported by DFG (German Research Foundation) Grant GE 3106/1-1. Flow cytometry analyses were performed in the Microbiology and Immunology Core Facility at Columbia University Medical Center. Qubit and Bioanalyzer tests for ATAC-seq were done at the Molecular Pathology/Molecular Pathology Shared Resource core of the Herbert Irving Comprehensive Cancer Center at Columbia University Medical Center (core-supporting NIH Grant P30CA013696).
Footnotes
Reviewers: K.B., The Francis Crick Institute; and H.R., Queen Mary University of London.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116735119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Coffin J. M., Hughes S. H., Varmus H. E., “The interactions of retroviruses and their hosts” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997). [PubMed] [Google Scholar]
- 2.Swanstrom R., Wills J. W., “Synthesis, assembly, and processing of viral proteins” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997). [PubMed] [Google Scholar]
- 3.Yoshimura F. K., Weinberg R. A., Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: Discovery of a smaller circular form. Cell 16, 323–332 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Kilzer J. M., et al. , Roles of host cell factors in circularization of retroviral DNA. Virology 314, 460–467 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Schwartzberg P., Colicelli J., Goff S. P., Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: A new viral function required for productive infection. Cell 37, 1043–1052 (1984). [DOI] [PubMed] [Google Scholar]
- 6.Sakai H., et al. , Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67, 1169–1174 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima N., Lu R., Engelman A., Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: Definition of permissive and nonpermissive T-cell lines. J. Virol. 75, 7944–7955 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon B., Chen I. S., Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77, 3962–3972 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G. Z., Wang Y., Goff S. P., Histones are rapidly loaded onto unintegrated retroviral DNAs soon after nuclear entry. Cell Host Microbe 20, 798–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y., Wang G. Z., Cingöz O., Goff S. P., NP220 mediates silencing of unintegrated retroviral DNA. Nature 564, 278–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geis F. K., Goff S. P., Unintegrated HIV-1 DNAs are loaded with core and linker histones and transcriptionally silenced. Proc. Natl. Acad. Sci. U.S.A. 116, 23735–23742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machida S., et al. , Exploring histone loading on HIV DNA reveals a dynamic nucleosome positioning between unintegrated and integrated viral genome. Proc. Natl. Acad. Sci. U.S.A. 117, 6822–6830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornberg R. D., Chromatin structure: A repeating unit of histones and DNA. Science 184, 868–871 (1974). [DOI] [PubMed] [Google Scholar]
- 14.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Elsässer S. J., Noh K. M., Diaz N., Allis C. D., Banaszynski L. A., Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf G., et al. , On the role of H3.3 in retroviral silencing. Nature 548, E1–E3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsässer S. J., Noh K. M., Diaz N., Allis C. D., Banaszynski L. A., Elsässer et al. reply. Nature 548, E7–E9 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Gurard-Levin Z. A., Quivy J. P., Almouzni G., Histone chaperones: Assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83, 487–517 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess R. J., Zhang Z., Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridgway P., Almouzni G., CAF-1 and the inheritance of chromatin states: At the crossroads of DNA replication and repair. J. Cell Sci. 113, 2647–2658 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Goh W. C., et al. , HIV-1 Vpr increases viral expression by manipulation of the cell cycle: A mechanism for selection of Vpr in vivo. Nat. Med. 4, 65–71 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Fregoso O. I., Emerman M., Activation of the DNA damage response is a conserved function of HIV-1 and HIV-2 Vpr that is independent of SLX4 recruitment. MBio 7, e01433-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J., Shun M. C., Zhang Y., Hao C., Skowronski J., HIV-1 Vpr counteracts HLTF-mediated restriction of HIV-1 infection in T cells. Proc. Natl. Acad. Sci. U.S.A. 116, 9568–9577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufman P. D., Kobayashi R., Kessler N., Stillman B., The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell 81, 1105–1114 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Serra-Cardona A., Zhang Z., Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem. Sci. 43, 136–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer P. V., et al. , Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 46, 9907–9917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazuda D. J., et al. , Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287, 646–650 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Wang J., et al. , RbAp48, a novel inhibitory factor that regulates the transcription of human immunodeficiency virus type 1. Int. J. Mol. Med. 38, 267–274 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Yang B. X., et al. , Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163, 230–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murzina N., Verreault A., Laue E., Stillman B., Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Lechner M. S., Schultz D. C., Negorev D., Maul G. G., Rauscher F. J. III, The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 331, 929–937 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Ma X., et al. , Histone chaperone CAF-1 promotes HIV-1 latency by leading the formation of phase-separated suppressive nuclear bodies. EMBO J. 40, e106632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geis F. K., Goff S. P., Silencing and transcriptional regulation of endogenous retroviruses: An overview. Viruses 12, 884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen T. S., et al. , Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Day D. S., Luquette L. J., Park P. J., Kharchenko P. V., Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 11, R69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf D., Goff S. P., TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Rowe H. M., et al. , KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Weinberg J. B., Matthews T. J., Cullen B. R., Malim M. H., Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174, 1477–1482 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis P., Hensel M., Emerman M., Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11, 3053–3058 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe T., Reynolds T. C., Yu G., Brown P. O., Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12, 2099–2108 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis P. F., Emerman M., Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68, 510–516 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zila V., et al. , Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupont L., et al. , The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr. Cell Host Microbe 29, 792–805.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon B., Chang M. A., Chen I. S., Vpr is required for efficient Nef expression from unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 81, 10515–10523 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon A. P., Gu H., Roizman B., ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 9993–9998 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merkl P. E., Orzalli M. H., Knipe D. M., Mechanisms of host IFI16, PML, and Daxx protein restriction of herpes simplex virus 1 replication. J. Virol. 92, e00057-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Washington S. D., et al. , The CCCTC binding factor, CTRL2, modulates heterochromatin deposition and the establishment of herpes simplex virus 1 latency in vivo. J. Virol. 93, e00415-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddi T. S., Merkl P. E., Lim S. Y., Letvin N. L., Knipe D. M., Tripartite motif 22 (TRIM22) protein restricts herpes simplex virus 1 by epigenetic silencing of viral immediate-early genes. PLoS Pathog. 17, e1009281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucifora J., Baumert T. F., Silencing of the HBV episome through degradation of HBx protein: Towards functional cure? J. Hepatol. 74, 497–499 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Irwan I. D., Karnowski H. L., Bogerd H. P., Tsai K., Cullen B. R., Reversal of epigenetic silencing allows robust HIV-1 replication in the absence of integrase function. MBio 11, e01038-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irwan I. D., Cullen B. R., Tax induces the recruitment of NF-κB to unintegrated HIV-1 DNA to rescue viral gene expression and replication. J. Virol. 95, e0028521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.