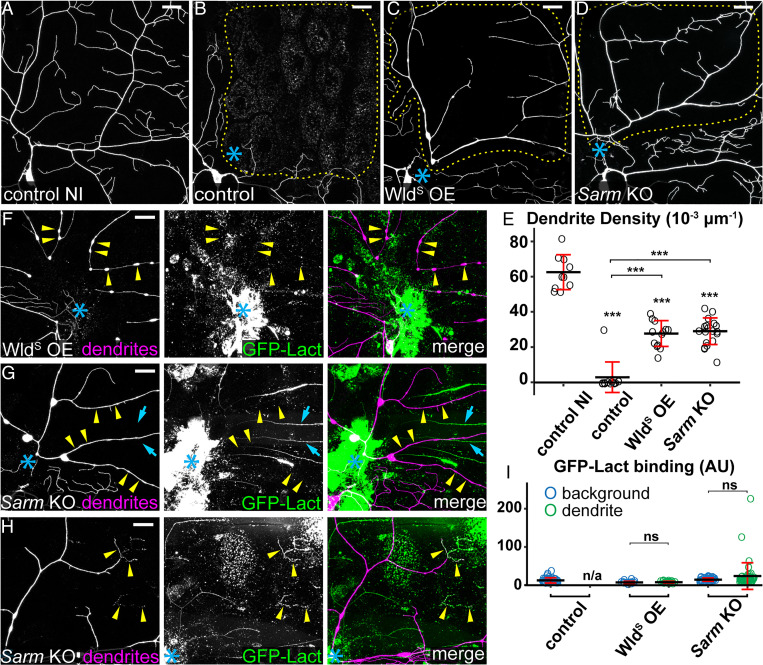
Fig. 3.
WldS OE and Sarm KO suppress PS exposure and fragmentation of injured dendrites. (A) Partial dendritic field of an uninjured ddaC control neuron. (B–D) Partial dendritic fields of control (B), WldS OE (C), and Sarm KO (D) ddaC neurons 24 h AI. Blue asterisk, injury site. Yellow dots outline regions covered by injured dendrites. (E) Quantification of dendrite density (1,000 × dendritic length/area) in regions covered by injured dendrites. n = number of neurons: control no injury (NI) (n = 10, 10 animals); control (n = 12, six animals); WldS OE (n = 13, six animals); and Sarm KO (n = 19, six animals). (F and G) WldS OE (F) and Sarm KO (G) ddaC neurons 24 h AI, showing unfragmented dendrites and the lack of GFP-Lact binding on severed dendrites (yellow arrowheads) of ddaC neurons. GFP signals that do not colocalize with tdTom signals are likely GFP-Lact binding to injured non-C4da neurons without Sarm KO (cyan arrows). (H) Dendrites of a Sarm KO ddaC neuron 24 h AI, showing infrequent GFP-Lact binding at fragmented terminal branches (yellow arrowheads). (I) Quantification of GFP-Lact binding on injured dendrites. The GFP-Lact intensity is shown for both background epidermal regions and injured dendrites. The outliers in Sarm KO dendrite dataset correspond to infrequent degenerating terminal branches. Background: epidermal regions without dendrites. n = number of measurements: control background (n = 39) and control dendrite (n/a, no remaining dendrites), seven animals; WldS OE background (n = 18) and WldS OE dendrite (n = 18), four animals; and Sarm KO background (n = 48) and Sarm KO dendrite (n = 48), nine animals. In all panels, neurons were labeled by ppk-MApHS (A–C; only tdTom channel is shown), ppk > CD4-tdTom (D), and ppk-CD4-tdTom (F–H). Neuronal-specific KO was induced by SOP-Cas9. For all quantifications, one-way ANOVA and Tukey test, ***P ≤ 0.001, ns, not significant, and n/a, not applicable. The significance level above each genotype in E is for comparison with the control. Black bar, mean; red bar, SD. (Scale bars, 25 μm.)