Fig. 3. Specificity and oligloclonality of lung CD4+ T-cells in LTA1/OmpX and OmpC vaccinated mice.
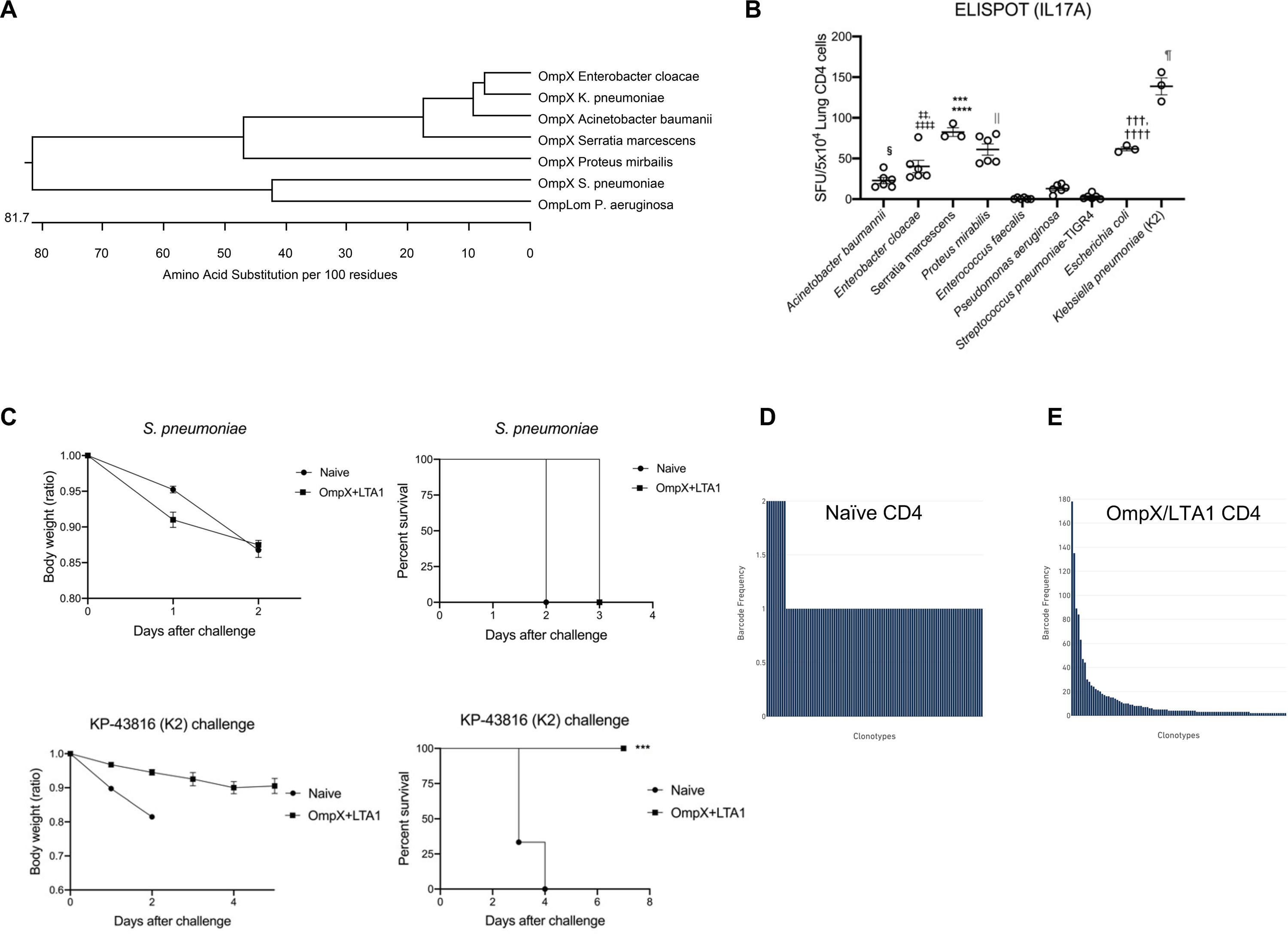
(A) Dendrogram analysis of OmpX protein sequences across members of the Enterobacteriaceae family, as well as the OmpLom orthologues in P. aeruginosa and S. pneumoniae. (B) Lung CD4+ T cells from OmpX+LTA1 immunized mice were isolated with MACS kit and stimulated with extracts of sonicated whole bacteria to enumerate IL-17A-producing cells by ELISpot. Significant differences were calculated by ANOVA followed by Tukey’s multiple comparisons test (n = 3–6, representative from 2 independent experiments). ****, P < 0.0001 (vs Acinetobacter, Enterococcus, Pseudomonas and Pneumococcus), ***, P < 0.001 (vs Enterobacter), ‡‡‡‡ P < 0.0001 (vs Enterococcus, Pseudomonas and Pneumococcus), ‡‡P < 0.01 (vs Acinetobacter), ††††P < 0.0001 (vs Enterococcus, Pseudomonas and Pneumococcus), †††P < 0.01 (vs Acinetobacter), §P < 0.05 (vs Enterococcus, Pneumococcus), || P < 0.0001 (vs Acinetobacter, Enterococcus, Pseudomonas and Pneumococcus), and ¶P < 0.0001 (vs all other bacteria). (C) Weight loss (as a ratio of starting weight) and survival in the naïve and OmpX+LTA1 vaccinated mice after challenge with 106 CFU of type 3 Streptococcus pneumoniae or KP-43816 (K2 strain) (n = 6, cumulative data of two separate experiments are shown). T cell clonotypes and single cell TCR seq for (D) naïve CD4+ T cells from spleen or (E) OmpX+LTA1 immunized lung CD4+ T cells were performed. Significant differences were estimated by one-way ANOVA followed by Tukey’s multiple comparisons test ***, P < 0.001.
