Figure 6. SCP4 functions upstream of STK35/PDIK1L to support AML proliferation.
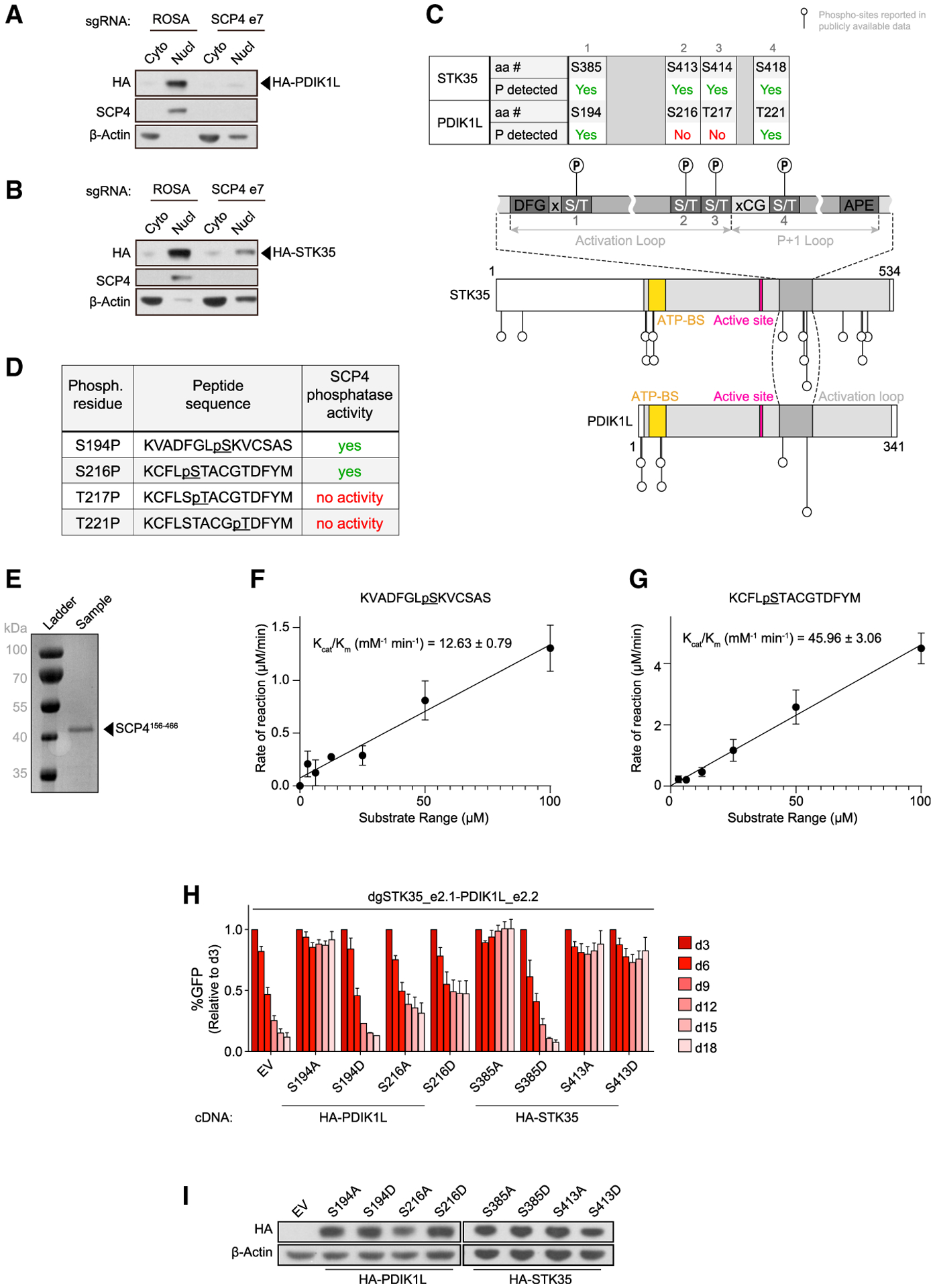
(A and B) Western blot of cytoplasm (Cyto) and nucleus (Nucl) fractions of MOLM-13 cells stably expressing HA-PDIK1L (A) or HA-STK35 (B) on day 5 post-infection with the indicated sgRNAs. Shown is a representative experiment of 3 independent biological replicates.
(C) Schematics of phosphorylation sites reported in the publicly available phospho-proteomics datasets on STK35 and PDK1L relative to their domain architectures (Hornbeck et al., 2015; Ochoa et al., 2020). aa #, amino acid number; P, phosphorylation; ATP-BS, ATP-binding site.
(D) Phosphorylated peptides assayed for SCP4 phosphatase activity in vitro.
(E) Recombinant SCP4 protein purity as assessed by SDS-PAGE and Coomassie blue staining. His-SUMO-(TEV)-SCP4 was expressed in BL21 (DE3) cells and purified by affinity, anion exchange, and gel filtration chromatography. Molecular weight markers are shown for reference.
(F and G) Phosphatase activity of SCP4 against the indicated peptides plotted for kinetic fitting. Each measurement was conducted in triplicate with standard deviations shown as error bars.
(H) Competition-based proliferation assay in MOLM-13 cells stably expressing EV or the CR HA-PDIK1L or HA-STK35 constructs harboring the indicated amino acid substitutions infected with the indicated sgRNAs. n = 3.
(I) Western blot of whole-cell lysates from MOLM-13 cells stably expressing EV or CR HA-PDIK1L or HA-STK35 constructs harboring the indicated amino acid substitutions.
All bar graphs represent the mean ± SEM. See also Figure S6.
