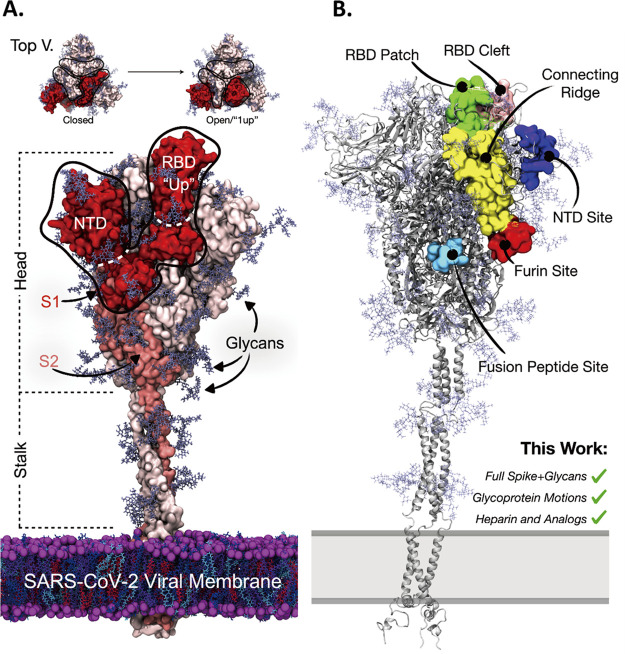
Figure 2.
(A) Molecular representation of SARS-CoV-2 spike in the “1-up” conformational state. The spike protein is represented with red, salmon, and light pink surfaces. Spike glycan atoms are shown with light blue licorice representation. (top) From a top-down view the “closed” to “open”/“1-up” RBD conformational change required for host-cell invasion. The spike’s S1 domain is highlighted in red surface representation, while the spike’s S2 domain is in salmon surface representation. The spike’s RBD and NTD are outlined for reference. (B) Molecular representation of the SARS-CoV-2 spike in closed conformation depicting literature proposed HEP binding sites. Green surface: the “RBD patch”, a site proposed by Skidmore and co-workers23 and supported by Esko and co-workers to have high affinity for heparin.27 Pink surface: the “RBD cleft” a site proposed by Fadda and co-workers to have high affinity for polysaccharides.34 Red surface: the furin cleavage site.37−39 Light blue surface: the fusion peptide site proposed by Linhardt et al.33 Yellow surface: the connecting ridge proposed by Wade et al.35 Purple surface: the NTD site identified by Schuurs et al.36