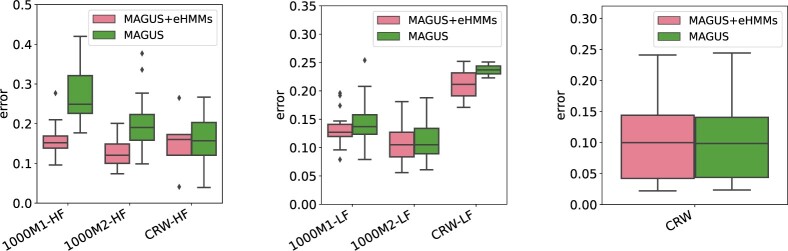
Fig. 1.
Experiment 2: Average alignment error in high fragmentation, low fragmentation, and without introduced fragmentation for MAGUS and MAGUS+eHMMs, from left to right. ‘CRW’ refers to datasets originated from Comparative Ribosomal Website (Cannone et al., 2002). ‘CRW-HF’ contains the high-fragmentation version of 16S.M, 23S.M (smaller datasets), 16S.3, 16S.T and 16S.B.ALL (larger datasets), ‘CRW-LF’ contains the low-fragmentation version of 16S.M and 23S.M, and ‘CRW’ contains the 14 CRW datasets without introduced fragmentation. For both high- and low-fragmentation conditions, we examine ROSE 1000M1, 1000M2 and CRW 16S.M, 23S.M datasets. For the high-fragmentation condition, we examine three additional large CRW datasets (16S.3, 16S.T and 16S.B.ALL with high fragmentation). For individual CRW dataset results with and without introduced fragmentation, see Supplementary Tables S1 and S3