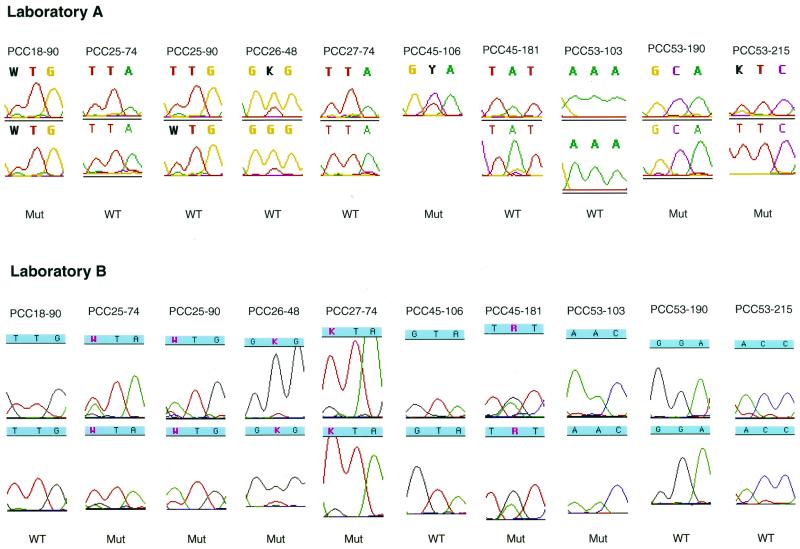
FIG. 5.
Chromatographic tracings showing the automated DNA sequence analysis of the 10 instances in which a primary resistance mutation was detected by just one laboratory (the PCC numbers at the tops of the tracings indicate patient code-codon number). In seven instances, the laboratory detecting the mutation detected it as part of a mixture, and in three instances (for patient PCC53) there was a complete mismatch between the two laboratories. The tracings for the forward and reverse sequences of each laboratory are shown (the reverse sequence at RT codon 106 for patient PCC45 at laboratory A is missing). Nucleotides with mixtures are shown in bold for laboratory A and in red for laboratory B. Laboratory A reported protease codon 90 for patient PCC25 and protease codon 48 for patient PCC26 as being of the wild type (WT) because the mutant (Mut) peak was detected in only one of the sequencing reactions. Laboratory A's reverse sequence of RT codon 181 for patient PCC45 shows a mixture of TAT and TGT. However, the minor G peak was not flagged by the ABI FACTURA program at the then-recommended cutoff of 30%. For three of the mixtures (for patient PCC25, codon 90; patient PCC45, codon 106; and patient PCC45, codon 181), the predominant nucleotide (having the larger peak) was different between the two laboratories.