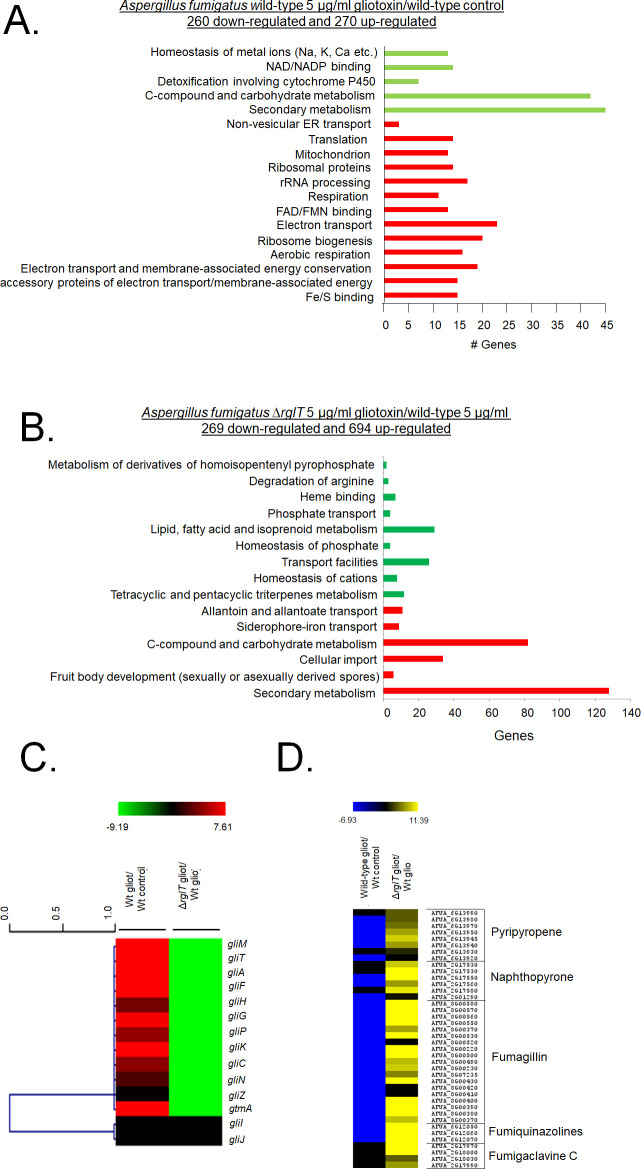
Fig 1. Functional characterisation (FunCat) of significantly differently expressed genes (DEGs) identified by RNA-sequencing in the A. fumigatus ΔrglT strain.
(A) FunCat analysis of DEGs up-regulated in the A. fumigatus wild-type and in (B) ΔrglT strain in comparison to the wild-type (WT) strains when exposed to 5 μg/ml GT for 3 h. (C) Heat map depicting the log2 fold change (Log2FC) of differentially expressed genes (DEGs), as determined by RNA-sequencing, and encoding enzymes present in the GT BGC required for GT biosynthesis. The gene gtmA was also included in this heat map. (D) Heat map depicting the Log2FC of differentially expressed genes (DEGs), as determined by RNA-sequencing, and encoding enzymes required for secondary metabolite (SM) biosynthesis. In both (C) and (D) log2FC values are based on the wild-type strain exposed to GT in comparison to the wild-type control and the ΔrglT exposed to GT in comparison to the wild-type exposed to GT. Heat map scale and gene identities are indicated. Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Pearson correlation with complete linkage clustering.