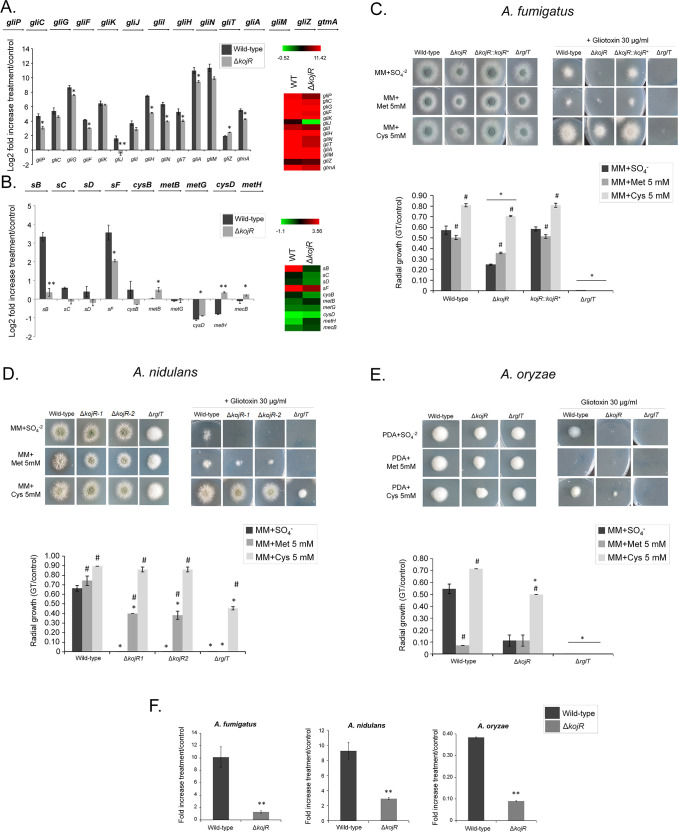
Fig 11. A. fumigatus gliJ and sulfur assimilation genes are dowregulated in the ΔkojR mutant.
– A. fumigatus was grown for 21 h at 37°C before 5 μg/ml of GT was added to cultures for 3 hours. RNA was extracted from the wild-type and ΔkojR strains, reverse transcribed to cDNA and RT-qPCRs were peformed. Results are expressed as log2 fold increase and are the average of three biological replicates ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (**, p < 0.01 and *, p < 0.005). (A) Genes in the A. fumigatus GT pathway and gtmA. (B) Selected genes in the A. fumigatus sulfur assimilation and transsulfuration pathways. Heat map scale and gene identities are indicated. (C) A. fumigatus, (D) A. nidulans, and (E) A. oryzae wild-type, ΔkojR, ΔkojR::kojR+, and ΔrglT mutants were grown for 48 hours at 37°C on MM+SO4-2, MM+Met 5 mM, and MM+Cys 5 mM in the presence or absence of 10 or 30 μg/ml of GT. The results are the average of three repetitions ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (*, p < 0.005 when comparing the mutant strains versus the wild-type and and #, p < 0.005 when comparing the growth on MM+Met or MM+Cys versus the growth on MM+SO4-). (F) A. fumigatus, A. nidulans, and A. oryzae were grown for 20 h at 37°C before 5 μg/ml of GT was added to cultures for 3 hours. RNA was extracted from the wild-type and ΔkojR strains, reverse transcribed to cDNA and RT-qPCRs were peformed. Results are the average of three biological replicates ± standard deviation. Statistical analysis was performed using a one-tailed, paired t-test when compared to the control condition (**, p < 0.01 and *, p < 0.005).