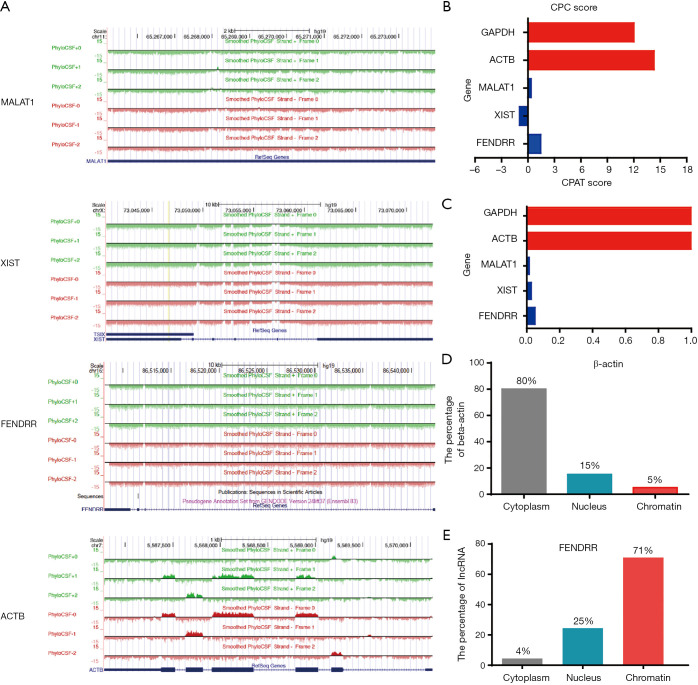
Figure 3.
The characteristics of lncRNA FENDRR in non-small cell lung cancer (NSCLC). (A) The coding potential of FENDRR was analyzed by the PhyloCSF CSF analysis. MALAT1 (metastasis associated lung adenocarcinoma transcript 1) and XIST (X inactive specific transcript) served as noncoding RNA controls, and ACTB served as coding RNA control. The regions with a score greater than 0 (above the baseline) were predicted to be coding, while regions with a score less than 0 (below the baseline) were predicted to be noncoding; (B,C) the coding potential of FENDRR was analyzed by Coding Potential Calculator (CPC) and coding potential Assessment Tool (CPAT). MALAT1 and XIST served as noncoding RNA controls, and ACTB and GAPDH served as coding RNA controls; (D,E) subcellular fractionation experiment to show the localization of FENDRR in cells.