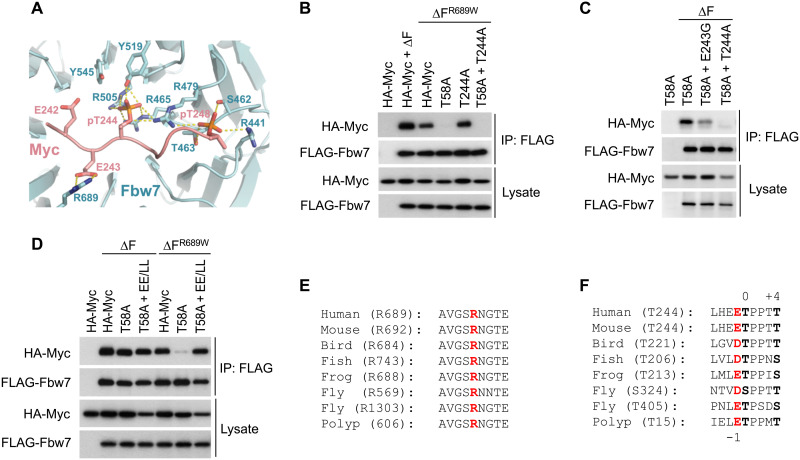
Fig. 2. Structure analysis of the T244 degron.
(A) Close-up view of the interface formed between Fbw7 and the Myc T244 degron. Fbw7 is colored in cyan with its Myc-interacting residues shown in sticks. The Myc T244 degron is colored in salmon. Hydrogen bonds and electrostatic interactions are indicated by yellow dashed lines. (B to D) HEK293 cells were transfected as indicated, immunoprecipitated against FLAG-Fbw7 (∆F), and blotted for associated Myc with HA antibody. (E) Alignment of the Fbw7 region centered at R689 (red). Bird, chicken; fish, zebrafish; frog, Xenopus; worm, Caenorhabditis elegans; fly, fruitfly; and polyp, Hydra vulgaris. Polyp Fbw7 was assembled from predicted partial transcripts. (F) Alignment of the T244 degron. The central 0 and +4 position phosphorylations are in black and bold, and the conserved negative charge in −1 is in red. Fruitflies lack the T58 degron entirely and contain two versions of the T244 degron instead, while hydra’s Fbw7 degrons are reversed such that the T244-type degron is located at the N terminus.