Abstract
Background
Platinum-based chemotherapy is the cornerstone of non-small cell lung cancer (NSCLC) therapy. However, the molecular mechanisms and predictive markers of platinum chemoresistance have not been fully understood. Our recent study revealed that Jumonji domain containing 5 (JMJD5) expression in cells was elevated under DNA damage by alkylating agent or UV radiation, which suggests a potential role of JMJD5 in DNA damage related chemoresistance. However, the role of JMJD5 in NSCLC chemotherapy has not been reported. In this study, we demonstrated JMJD5 as a potential prognostic indicator in NSCLC patients who received platinum-based chemotherapy.
Methods
JMJD5 protein expression level in tumor and adjacent normal tissues were detected by immunohistochemistry. Samples were from primary NSCLC patients who received platinum-based chemotherapy after surgical resection. Survival curves were presented by the Kaplan-Meier method and p value was acquired by log-rank test. Multivariate analysis was tested by Cox proportional-hazards regression method.
Results
Elevated JMJD5 expression was found in 27.2% cases of tumor tissues (22/81), and high JMJD5 expression were significantly associated with poor overall survival time (OS) [HR =2.881 (1.774–9.121), P=0.001] and progression-free survival time (PFS) [HR =2.255 (1.417–5.886), P=0.004] in NSCLC patients who received platinum-based chemotherapy. In multivariate analyses, JMJD5 was proved to be an independent prognostic indictor for shorter OS [HR =2.339 (1.158–4.724), P=0.018] and PFS [HR =2.031 (1.095–3.767), P=0.025).
Conclusions
High JMJD5 expression indicated a worse prognosis in NSCLC patients who received platinum-based chemotherapy, and JMJD5 may serve as a novel predictive marker in NSCLC chemotherapy.
Keywords: Jumonji domain containing 5 (JMJD5), prognosis, non-small cell lung cancer (NSCLC), platinum-based chemotherapy, survival outcome
Introduction
One-quarter of all cancer deaths are due to lung cancer, while non-small cell lung cancer (NSCLC) accounts for 85–90% of total pulmonary malignancies (1). Although great progress has been made in targeted therapy and immunotherapy, the platinum-based chemotherapy is still the cornerstone of NSCLC treatment. However, chemoresistance has never been overcome and molecular indicators are urgently needed for prediction of chemotherapy response. Previous studies have shown that DNA damage repair is an important pathway leading to platinum-based chemotherapy resistance (2). Recently, our group reported that Jumonji domain containing 5 (JMJD5) regulates gene transcription through clipping of monomethylated histone H3 N-tail under DNA damaging stress (3), which suggested a potential role of JMJD5 in chemoresistance.
JMJD5, also known as KDM8, was reported to be responsible for gene transcription regulation through its enzymatic activity towards H3K36me2 (4-6) and to regulate osteoclastogenesis with its hydroxylase activity (7). Despite the controversy exists on its demethylase activity (8,9), JMJD5 plays a crucial role in embryonic development (4,10,11), circadian rhythms (12), cell-cycle progression (13-15), and also in tumor genesis (5,16-20). Down-regulation of JMJD5 suppresses metastasis and induces apoptosis in oral squamous cell carcinoma by regulating p53/NF-κB pathway (20). JMJD5 was also reported as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in castration-resistant prostate cancer (21). Chromatin immunoprecipitation assays revealed that JMJD5 regulated cyclin A1 transcription through directly remove the methyl group of H3K36me2 on its coding region and thus promoted cancer cell proliferation (5). JMJD5 was also found interacted with p53 DNA-binding domain (DBD) and negatively regulated its activities in cell cycle and proliferation (14). Additionally, JMJD5 interacts with pyruvate kinase muscle isozyme (PKM)2 and regulates its nuclear translocation. The interaction between JMJD5 and PKM2 promotes hypoxia inducible factor (HIF)-1α-mediated glucose metabolism, a proverbial metabolic way which generates energy for cancer cells to meet their rapid growth demands (22). This might explain why JMJD5 was found overexpressed in several types of cancer (5) and was positively related to a poor survival outcome in breast cancer (17) and colon cancer (16).
Recent study reported that JMJD5 functioned in mitosis progression and depletion of JMJD5 resulted in significant mitotic arrest and spindle assembly defects (15). The same team also demonstrated that depletion of JMJD5 sensitized tumor cells to microtubule-destabilizing agents, such as vincristine and colchicine, by altering microtubule stability and promoted tumor cell apoptosis (18). In a word, all those results implicated JMJD5 as a potential oncogene. However, in hepatocellular carcinoma (HCC) pathogenesis, JMJD5 seems to be a tumor suppressor gene, since silence of JMJD5 down regulates CDKN1A transcription and promotes HCC cell proliferation (19). The diverse outcomes implicated JMJD5 may play inverse roles in the context of different carcinoma types. To our knowledge, the role of JMJD5 expression in NSCLC has not been demonstrated. In this study, we illustrated JMJD5 as a predictive marker which indicated a poor survival outcome in NSCLC patients who received platinum-based chemotherapy.
Methods
Patients and samples
In this retrospective study, NSCLC patients who received platinum-based chemotherapy after surgery at Zhejiang Cancer Hospital (Hangzhou, China) between July 2011 and October 2012 were screened. Only patients with primary NSCLC tumor were included. This study was approved by the Ethics Committee of Zhejiang Cancer Hospital. Tumor tissues were collected from the department of tumor tissue bank of Zhejiang Cancer Hospital, China. TNM stages were determined according to the 7th edition of the international staging system (23). Computed tomography (CT), ultrasonography, magnetic resonance imaging (MRI), and chest radiography were reviewed to evaluate disease recurrence status. Follow-up work was done through telephone. The ultimate follow-up time was up to May 20th, 2019.
Immunohistochemical (IHC) staining and evaluation
JMJD5 expression in both tumor and normal tissues of the same lung cancer samples were determined by IHC staining. In brief, formalin fixed paraffin-embedded (FFPE) tumor tissues were cut into a series of 5µm thick sections. The sections were de-paraffinized and rehydrated. Antigen retrieval was achieved using Dako EnVisionTM FLEX Target Retrieval Solution (pH 9.0) in the Dako PT Link units at 95 °C for 20 minutes. Thereafter, each section was incubated with the anti-JMJD5 antibody in 1:100 (Rabbit polyclonal antibody, ab83011, Abcam) for 2 hours at room temperature. Followed by incubation with secondary antibody (PV-9000, Zhongshan Jinqiao Biotechnology Co. Ltd., Beijing, China) at room temperature for 30 minutes. Finally, the sections were colorized by 3,3'-diaminobenzamine and hematoxylin, respectively for immune complex and nuclei staining.
Both staining intensity and the percentage of positive tumor cells were evaluated as follows: (I) staining intensity: 0, absence of staining; 1, weak staining; 2, moderate staining; and 3, strong staining; (II) percentage of positive tumor cells: 0, 0–25% of staining; 1, 25–50% staining; 2, 50–75% staining; 3, ≥75% staining. The final score was obtained by multiplying the staining intensity score and percentage score. Final score ≥6 was defined as high expression.
Statistical analyses
The SPSS software for Windows (Version 17.0, SPSS, Inc., Chicago, IL, USA) was used for statistical analyses. Overall survival time (OS) and progression-free survival time (PFS) were defined as the months between surgery date to death date or disease progression date, respectively. Survival curves were presented by the Kaplan-Meier method and P value was acquired using the log-rank test. Multivariate analysis for JMJD5 and other potential prognostic variables (including gender, TNM stage, tumor differentiation status) was tested by Cox proportional-hazards regression method. Chi-squared (χ2) and Fisher’s exact tests were used to evaluate the baseline of JMJD5 expression in different clinicopathological groups. For all tests, P values were two-sided and P<0.05 was considered statistically significant.
Results
Patients
Finally, a total of 81 patients who received platinum-based chemotherapy were included in this study. Baseline characteristics were showed in Table 1. The median of age is 59 [40–78]. Most of patients were male (80.2%), at early stage (stage I & II, 65.4%) or with smoking history (75.3%). During an 8-year follow-up, the median OS is 48.5 months (3.6–94.2 months). Thirty-six patients (44.4%) died, and 51 patients (63.0%) have disease progression during the period of follow-up.
Table 1. Clinicopathologic characteristics of overall patients (n=81).
| Characteristics | Subgroup | Patients, n (%) |
|---|---|---|
| Age (y) | <60 | 43 (53.1) |
| ≥60 | 38 (46.9) | |
| Gender | Male | 65 (80.2) |
| Female | 16 (19.8) | |
| Histopathology | Adenocarcinoma | 36 (44.4) |
| Squama | 43 (53.1) | |
| Othersa | 2 (2.5) | |
| Lymph node metastasis | No | 30 (37.0) |
| Yes | 51 (63.0) | |
| TNM stage | I–II | 53 (65.4) |
| III–IV | 28 (34.6) | |
| Differentiation | Poor | 45 (55.6) |
| Good-moderate | 36 (44.4) | |
| Smoking history | No | 20 (24.7) |
| Yes | 61 (75.3) | |
| JMJD5 expression status | Low | 59 (72.8) |
| High | 22 (27.2) |
a, others including adenosquamous carcinoma, large cell carcinoma, sarcoma, etc.
JMJD5 expression is elevated in NSCLC tumor tissues
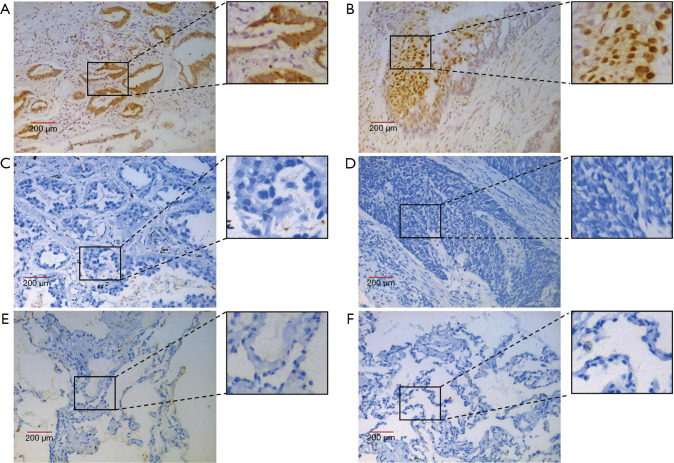
The JMJD5 staining was mainly localized in the nuclei and rare in the cytoplasm. As described above, we define high and low JMJD5 expression groups according to its staining intensity and positive percentage. Twenty-two/81 (27.2%) cases were classified as high JMJD5 expression. Figure 1A,B showed high JMJD5 respectively in adenocarcinoma tumor and squamous carcinoma, while Figure 1C,D showed low/negative JMJD5 expression in NSCLC tumors. In adjacent non-tumor lung tissues from NSCLC patients, a few cells were detected with JMJD5 expression (Figure 1E,F). JMJD5 expression in patients with different clinicopathological characteristics were showed in Table 2. JMJD5 expression showed no significant bias in different subgroups of clinicopathological characteristics.
Figure 1.
JMJD5 expression in tumor cells and adjacent lung tissue. (A,B) High JMJD5 expression in adenocarcinoma and squamous carcinoma, which predominantly localized to the nuclear region; (C,D) low/negative JMJD5 expression in adenocarcinoma and squamous carcinoma; (E,F) a few cells have weak nuclear expression of JMJD5 in adjacent lung tissues. JMJD5, Jumonji domain containing 5.
Table 2. JMJD5 expression in different clinicopathological characteristics groups.
| Characteristics | JMJD5 | P value | |
|---|---|---|---|
| Low, n (%) | High, n (%) | ||
| Age (y) | 0.872 | ||
| <60 | 31 (72.1) | 12 (27.9) | |
| ≥60 | 28 (73.7) | 10 (26.3) | |
| Gender | 0.299 | ||
| Male | 49 (75.4) | 16 (24.6) | |
| Female | 10 (62.5) | 6 (37.5) | |
| Histopathology | 0.356 | ||
| Adenocarcinoma | 24 (66.7) | 12 (33.3) | |
| Squama | 34 (79.1) | 9 (20.9) | |
| Othersa | 1 (50.0) | 1 (50.0) | |
| Lymph node metastasis | 0.103 | ||
| No | 25 (83.3) | 5 (16.7) | |
| Yes | 34 (66.7) | 17 (33.3) | |
| TNM stage | 0.075 | ||
| I–II | 42 (79.2) | 11 (20.8) | |
| III–IV | 17 (60.7) | 11 (39.3) | |
| Differentiation | 0.371 | ||
| Poor | 31 (68.9) | 14 (31.1) | |
| Good-moderate | 28 (77.8) | 8 (22.2) | |
| Smoking history | 0.364 | ||
| No | 13 (65.0) | 7 (35.0) | |
| Yes | 46 (75.4) | 15 (24.6) | |
a, others including adenosquamous carcinoma, large cell carcinoma, sarcoma, etc. Data were analyzed by chi-square test. JMJD5, Jumonji domain containing 5.
High JMJD5 expression predicts shorter OS and PFS of NSCLC patients
We then evaluated the correlation between JMJD5 expression and patients’ survival outcomes using Kaplan-Meier method. Results showed in Figure 2 revealed that high JMJD5 expression indicated both shorter OS [HR =2.881 (1.774–9.121), P=0.001] and PFS [HR =2.255 (1.417–5.886), P=0.004]. To further elucidate whether JMJD5 is an independent prognostic factor, univariate and multivariate analysis were performed by Cox proportional hazards analyses and results are showed in Table 3. For univariate analysis, besides JMJD5, gender, histopathology and TNM stage were also indicators for shorter PFS, while only TNM stage was another indicator for shorter OS. For multivariate analysis, only JMJD5 (for PFS, P=0.025; for OS, P=0.018) and advanced disease stage (for PFS, P=0.007; for OS, P=0.017) were independent prognostic factors for PFS and OS.
Figure 2.
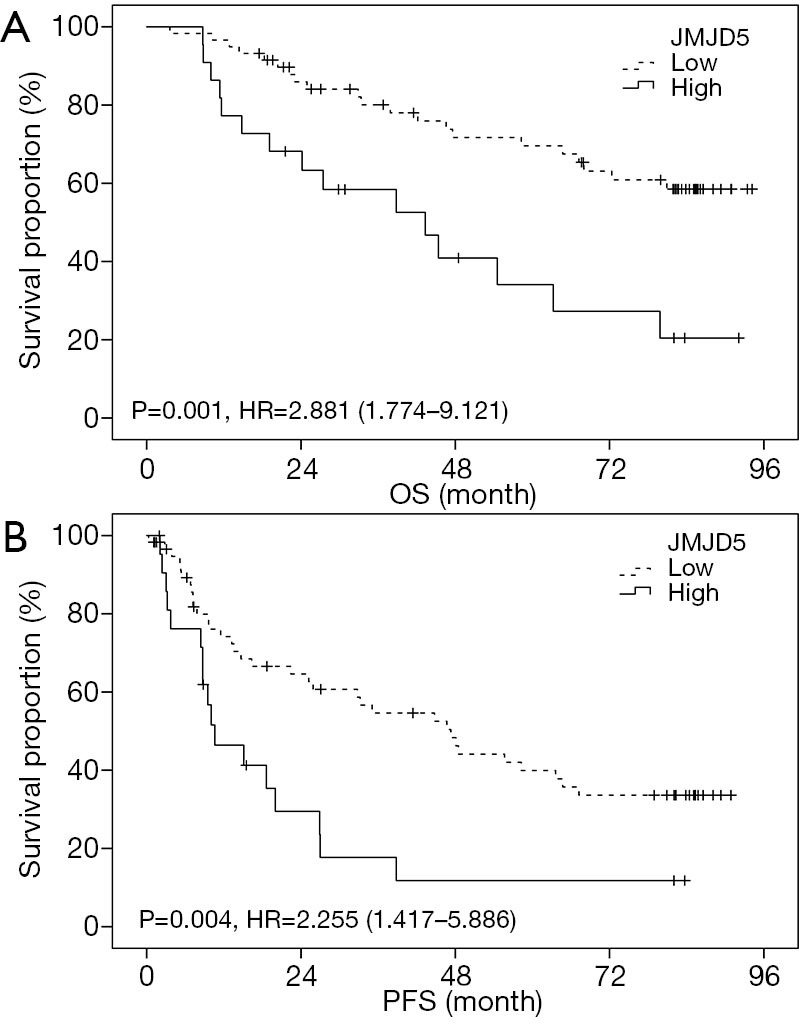
High JMJD5 expression predicts a poor OS and PFS. The survival curves were described by Kaplan-Meier analysis. High JMJD5 expression was associated with a shorter OS [HR =2.881 (1.774–9.121), P=0.001] and PFS [HR =2.255 (1.417–5.886), P=0.004]. The statistical significance was assessed by the log-rank test. JMJD5, Jumonji domain containing 5; OS, overall survival; PFS, progression-free survival.
Table 3. Univariate and multivariate Cox proportional hazards analyses for disease-free survival and overall survival.
| Variable | Progression-free survival | Overall survival | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Univariate | |||||
| Age (y) | |||||
| <60 | 1 | 1 | |||
| ≥60 | 0.868 (0.499–1.508) | 0.615 | 0.893 (0.463–1.724) | 0.737 | |
| Gender | |||||
| Male | 1 | 1 | |||
| Female | 1.917 (1.013–3.629) | 0.046* | 1.091 (0.497–2.395) | 0.829 | |
| Histopathology | |||||
| Adenocarcinoma | 1 | 1 | |||
| Squama | 1.782 (0.241–13.193) | 0.571 | 0.791 (0.104–5.994) | 0.821 | |
| Othersa | 0.597 (0.078–4.564) | 0.619 | 0.547 (0.072–4.177) | 0.561 | |
| Lymph node metastasis | |||||
| No | 1 | 1 | |||
| Yes | 1.028 (0.574–1.841) | 0.926 | 1.407 (0.678–2.922) | 0.359 | |
| TNM stage | |||||
| I–II | 1 | 1 | |||
| III–IV | 2.662 (1.523–4.652) | 0.001* | 2.846 (1.476–5.488) | 0.002* | |
| Differentiation group | |||||
| Poor | 1 | 1 | |||
| Good-moderate | 0.625 (0.386–1.010) | 0.055 | 0.732 (0.412–1.302) | 0.289 | |
| Smoking history | |||||
| No | 1 | 1 | |||
| Yes | 0.496 (0.272–0.905) | 0.496 | 0.942 (0.443–2.005) | 0.877 | |
| JMJD5 | |||||
| Low | 1 | 1 | |||
| High | 2.340 (1.286–4.257) | 0.005* | 2.950 (1.507–5.776) | 0.002* | |
| Multivariate | |||||
| Gender | |||||
| Male | 1 | 1 | |||
| Female | 1.624 (0.850–3.105) | 0.142 | 0.921 (0.416–2.039) | 0.839 | |
| TNM stage | |||||
| I–II | 1 | 1 | |||
| III–IV | 2.210 (1.244–3.926) | 0.007 | 2.324 (1.163–4.646) | 0.017* | |
| Differentiation group | |||||
| Poor | 1 | 1 | |||
| Good-moderate | 0.728 (0.437–1.212) | 0.222 | 0.938 (0.506–1.737) | 0.838 | |
| JMJD5 | |||||
| Low | 1 | 1 | |||
| High | 2.031 (1.095–3.767) | 0.025 | 2.339 (1.158–4.724) | 0.018* | |
a, others including adenosquamous carcinoma, large cell carcinoma, sarcoma, etc. Data were analyzed by chi-square test. Univariate and multivariate analysis was done using the Cox proportional hazard regression analysis. *, P<0.05. CI, confidence interval.
The predictive role of JMJD5 is the same in different subgroup
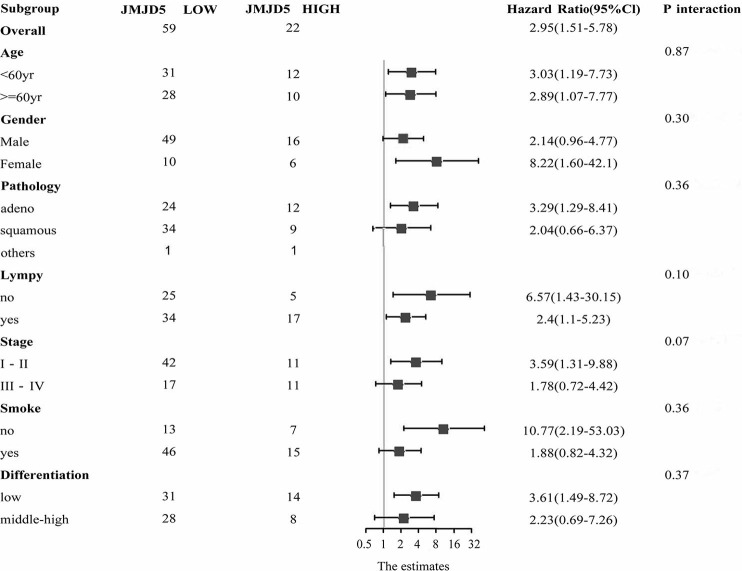
The results above implicated JMJD5 as an oncogene and a potential predictive biomarker for NSCLC. To further clarify the advantage population for JMJD5 to predict outcomes, a series of subgroup analyses were performed according to patients’ clinicopathological characteristics including age, gender, pathology, differentiation status, lymphatic metastasis, tumor stages and smoking history. Results were shown in Figure 3. No significant difference in HR were found in subgroup analyses (all P values are >0.05), which suggested there was no significant difference in the predictive effect of JMJD5 in each subgroup.
Figure 3.
Subgroup analyses HR for JMJD5 in predicting PFS and OS. No significant difference in HR were found in subgroup analyses (all P values are >0.05). JMJD5, Jumonji domain containing 5; OS, overall survival; PFS, progression-free survival.
Discussion
In this study, we reported higher JMJD5 expression indicating poor OS and PFS in NSCLC patients who received platinum-based chemotherapy. However, its molecular mechanism has not been well demonstrated. Our previous study revealed that JMJD5 regulates gene transcription via proteolytically clips monomethylated histone H3 under DNA damage stress. This could be one of the pathways which lead to chemoresistance. Despite the controversy on the epigenetic function of JMJD5 as demethylase or hydroxylase (8,9), important roles of JMJD5 in tumorigenesis and progression have already been implicated (5,16-19). Besides, its regulatory capacity including cell cycle regulation (14,15), glucose metabolism (21), and p53 regulation (14,24), all support its potential to promote cancer cell proliferation (5). In breast cancer (17,20) and colon cancer (16), JMJD5 expression was suggested as a worse prognostic factor. However, results in HCC pathogenesis was reversed, JMJD5 seemed to be a tumor suppressor gene (19). It may implicate that JMJD5 plays complicated roles in the context of different carcinoma types, while the role of JMJD5 in NSCLC is remain unclear. He et al. reported that JMJD5 interacted with spindle microtubules (15) and depletion of JMJD5 sensitized tumor cells to microtubule-destabilizing agents like vincristine (18), suggested a potential role of JMJD5 in chemoresistance. In this study, we demonstrated the predicting role of JMJD5 in survival outcome of chemotherapy in NSCLC.
No baseline differences of JMJD5 expression were found in subpopulation with various clinicopathologic characters. Survival analysis showed high JMJD5 expression was associated with a shorter OS and PFS. Multivariate analysis recognized that JMJD5 was an independent prognostic factor for both PFS and OS. In addition, subgroup analyses showed that no significant differences were found in the predictive effect of JMJD5 in subgroup population.
Moreover, recent study showed that JMJD5 was associated with the chemosensitivities of microtubule-targeting agents (18). Since navelbine and paclitaxel, the typical microtubule-targeting agents, were routinely used with platinum as first-line chemotherapy recommendation for NSCLC treatment, we are interested in whether the association between JMJD5 and chemosensitivities varies within patients received different chemotherapy regimens. However, as the sample size was limited, subgroup analysis with chemotherapy regimens was failed to be carried out in this study but would be illustrated in our following study.
Conclusions
JMJD5 was implied to be a prognostic marker for NSCLC patients who received platinum-based chemotherapy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81803549 to Xueping Xiang, 81702401 to Hong Liu and 81602465 to Hui Liu), the Natural Science Foundation of Zhejiang Province (LQ18H310002 to Xueping Xiang and LQ16H160014 to Hong Liu), and Projects of Medical and Health Technology Program in Zhejiang Province (2018KY026 to Yinghui Tong, 2015KYB061 and 2016KYB033 to Like Zhong).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Zhejiang Cancer Hospital, and was exempted from informed consent.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.16). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Cohen SM, Lippard SJ. Cisplatin: from DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol 2001;67:93-130. 10.1016/S0079-6603(01)67026-0 [DOI] [PubMed] [Google Scholar]
- 3.Shen J, Xiang X, Chen L, et al. JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep 2017;18:2131-43. 10.15252/embr.201743892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishimura A, Minehata K, Terashima M, et al. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 2012;139:749-59. 10.1242/dev.074138 [DOI] [PubMed] [Google Scholar]
- 5.Hsia DA, Tepper CG, Pochampalli MR, et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci U S A 2010;107:9671-6. 10.1073/pnas.1000401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuhrmann D, Mernberger M, Nist A, et al. Miz1 Controls Schwann Cell Proliferation via H3K36(me2) Demethylase Kdm8 to Prevent Peripheral Nerve Demyelination. J Neurosci 2018;38:858-77. 10.1523/JNEUROSCI.0843-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29. 10.3322/caac.20138 [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Zhou X, Wu M, et al. Structure of the JmjC-domain-containing protein JMJD5. Acta Crystallogr D Biol Crystallogr 2013;69:1911-20. 10.1107/S0907444913016600 [DOI] [PubMed] [Google Scholar]
- 9.Wilkins SE, Islam MS, Gannon JM, et al. JMJD5 is a human arginyl C-3 hydroxylase. Nat Commun 2018;9:1180. 10.1038/s41467-018-03410-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh S, Janknecht R. Histone demethylase JMJD5 is essential for embryonic development. Biochem Biophys Res Commun 2012;420:61-5. 10.1016/j.bbrc.2012.02.115 [DOI] [PubMed] [Google Scholar]
- 11.Ishimura A, Terashima M, Tange S, et al. Jmjd5 functions as a regulator of p53 signaling during mouse embryogenesis. Cell Tissue Res 2016;363:723-33. 10.1007/s00441-015-2276-7 [DOI] [PubMed] [Google Scholar]
- 12.Jones MA, Covington MF, DiTacchio L, et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A 2010;107:21623-8. 10.1073/pnas.1014204108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Hu S, Baker J. JMJD5 regulates cell cycle and pluripotency in human embryonic stem cells. Stem Cells 2014;32:2098-110. 10.1002/stem.1724 [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Zhang S, Qi H, et al. JMJD5 interacts with p53 and negatively regulates p53 function in control of cell cycle and proliferation. Biochim Biophys Acta 2015;1853:2286-95. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Wu J, Su X, et al. JMJD5 (Jumonji Domain-containing 5) Associates with Spindle Microtubules and Is Required for Proper Mitosis. J Biol Chem 2016;291:4684-97. 10.1074/jbc.M115.672642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Huang Q, Li Y, et al. JMJD5 is a potential oncogene for colon carcinogenesis. Int J Clin Exp Pathol 2015;8:6482-9. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Sun C, Li F, et al. Overexpression of histone demethylase JMJD5 promotes metastasis and indicates a poor prognosis in breast cancer. Int J Clin Exp Pathol 2015;8:10325-34. [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, He Z, Wang DL, et al. Depletion of JMJD5 sensitizes tumor cells to microtubule-destabilizing agents by altering microtubule stability. Cell Cycle 2016;15:2980-91. 10.1080/15384101.2016.1234548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu BH, Chen H, Cai CM, et al. Epigenetic silencing of JMJD5 promotes the proliferation of hepatocellular carcinoma cells by down-regulating the transcription of CDKN1A 686. Oncotarget 2016;7:6847-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Liu L, Ji F, et al. Soybean (Glycine max) prevents the progression of breast cancer cells by downregulating the level of histone demethylase JMJD5. J Cancer Res Ther 2018;14:S609-15. 10.4103/0973-1482.187292 [DOI] [PubMed] [Google Scholar]
- 21.Wang HJ, Pochampalli M, Wang LY, et al. KDM8/JMJD5 as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in CRPC. Oncogene 2019;38:17-32. 10.1038/s41388-018-0414-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HJ, Hsieh YJ, Cheng WC, et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1alpha-mediated glucose metabolism. Proc Natl Acad Sci U S A 2014;111:279-84. 10.1073/pnas.1311249111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobin LH, Gospodarowicz MK, Wittekind Ch. International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. Oxford, UK: Wiley-Blackwell; 2009. [Google Scholar]
- 24.Yao Y, Zhou WY, He RX. Down-regulation of JMJD5 suppresses metastasis and induces apoptosis in oral squamous cell carcinoma by regulating p53/NF-kappaB pathway. Biomed Pharmacother 2019;109:1994-2004. 10.1016/j.biopha.2018.07.144 [DOI] [PubMed] [Google Scholar]