Figure 2.
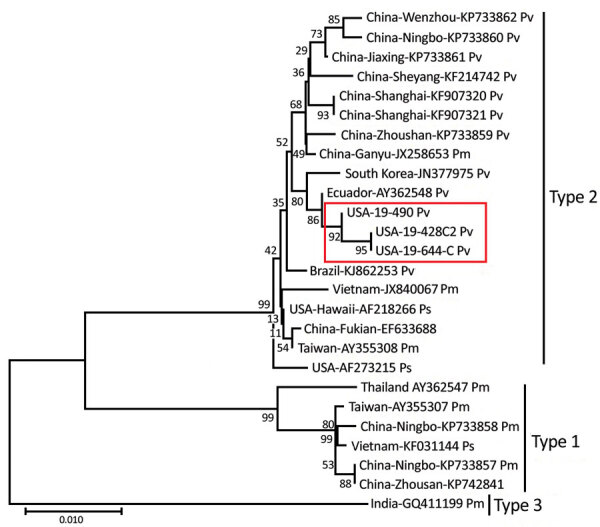
Evolutionary relationships of the infectious hypodermal and hematopoietic necrosis virus (IHHNV) strains (19-428, 19-490, and 19-644) recently detected in the United States and published capsid protein gene sequences. The recent IHHNV strains (red box) fall into the type 2 lineage. The evolutionary history was inferred by using the neighbor-joining method (24). The optimal tree with the sum of branch length = 0.20086053 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the maximum-likelihood method (25). Based upon full-genome phylogenetic analysis, the Texas and Florida IHHNV viruses appear to be related to a strain from Ecuador (GenBank accession no. AY362548.1). Scale bar indicates substitutions per site.
