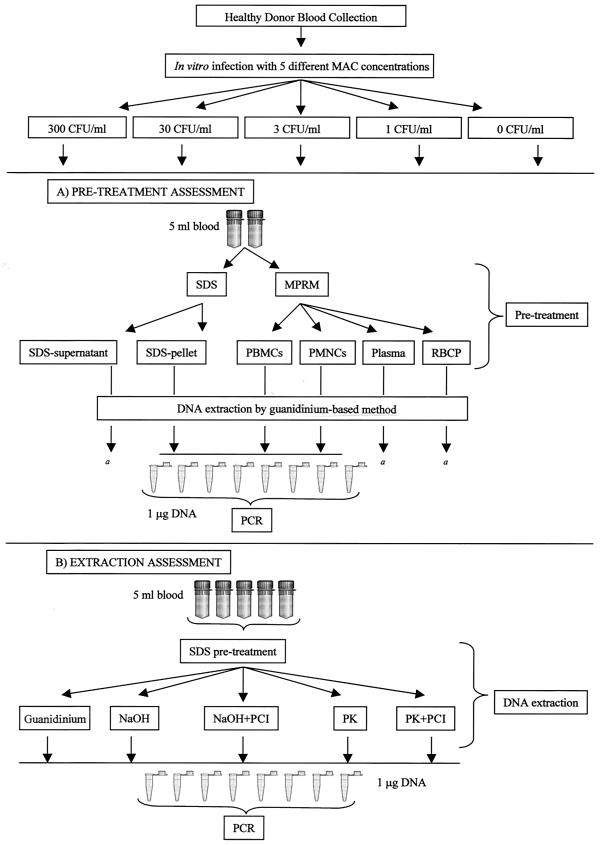
FIG. 1.
(A) Whole peripheral blood (50 ml) was divided into five parts (10 ml each). Four parts were inoculated in vitro with four different M. avium bacterial loads (300, 30, 3, and 1 CFU/ml), and the remaining part was utilized as a negative control. Each part was further subdivided to obtain two aliquots (5 ml each). These aliquots underwent one of two different pretreatments (lysis by SDS or cell separation by gradient centrifugation) to obtain three separate blood components: SDS-lysate pellet, PBMCs, and PMNCs. The guanidine-based method was used for DNA extraction. PCR amplification was performed with 1 μg of DNA per replicate, and eight replicates were amplified for each DNA sample. a, the presence of M. avium in SDS-lysate supernatants, plasma layers, and red blood cell pellets (RBCP) (after M-PRM separation) from 300-CFU/ml blood samples was also investigated; PCR analysis was performed for the totality of DNA extracted from these components. (B) Whole peripheral blood (125 ml) was divided into five parts (25 ml each). Four parts were inoculated in vitro with four different M. avium bacterial loads, and the remaining part was utilized as a negative control. Each part was further subdivided to obtain five aliquots (5 ml each). These aliquots underwent pretreatment with SDS. Three DNA extraction methods (guanidine-based method, NaOH-based method with or without purification, and PK-based method with or without purification) were used for DNA extraction. PCR amplification was performed with 1 μg of DNA per replicate, and eight replicates were amplified for each DNA sample. PCI, phenol-chloroform-isoamylalcohol.