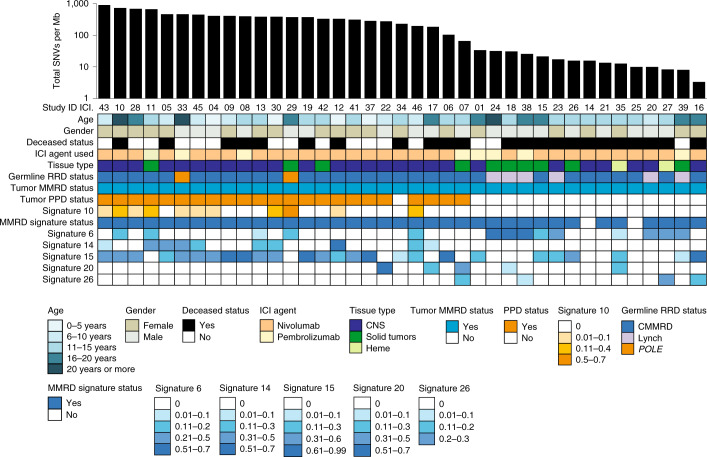
Fig. 3. Onco-plot summarising the genomic features from 39 available paired tumors and germline exomes, and their clinical correlates.
The tumors are arranged in descending order by total single nucleotide variants (SNVs) per megabase using semi-logarithmic scale. Tumor status for mismatch repair deficiency (MMRD) was determined based on MMR gene mutation status by whole exome sequencing (WES), or genetic testing of the patient germline. Tumor polymerase-proofreading deficiency (PPD) status was ascertained by presence of known POLE/POLD1 driver mutations and/or COSMIC signature status for PPD (COSMIC signature 10). The relative weight of each mutational signature contributing to each specific tumor sample analysis as reported using the DeconstructSigs package (Methods) has been color-coded, with the values provided below the onco-plot.