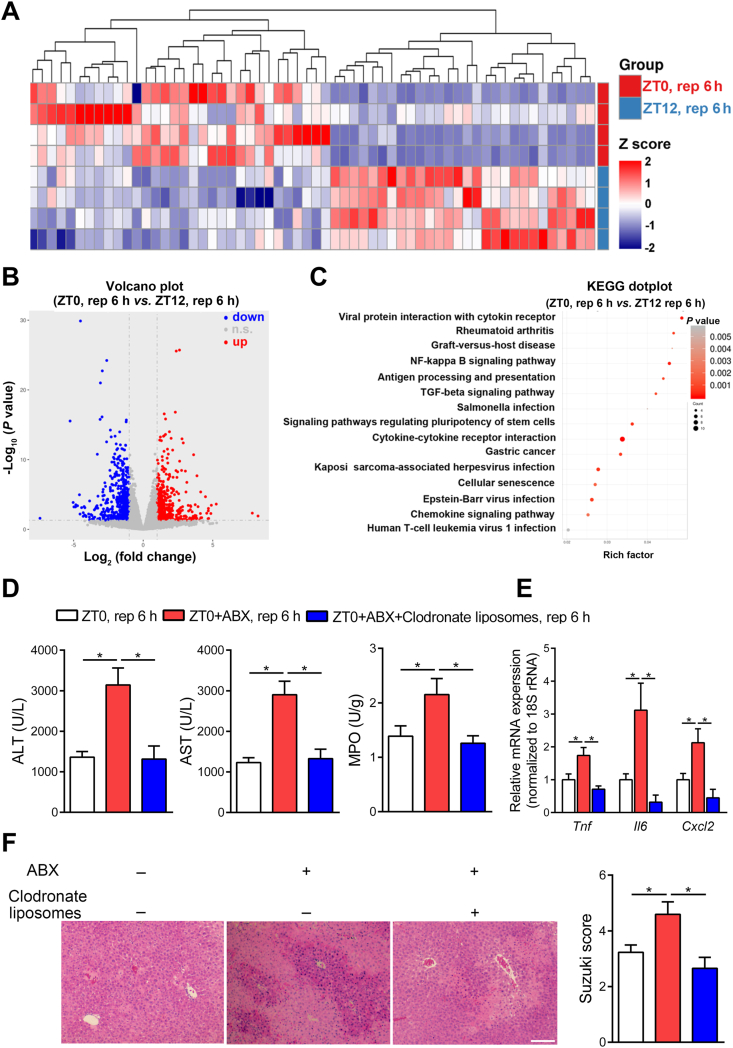
Figure 3.
Macrophages mediate the diurnal variation of HIRI in mice. (A–C) The differences between HIRI mice livers at ZT0 and ZT12 were analyzed by transcriptome. The heat map was performed to display the transcriptional level which was significant differentially expressed of inflammation-related genes in HIRI mice livers at ZT0 and ZT12. Red represents high relative expression and blue represents low relative expression (n = 4, A); The volcano plots of gene expression in HIRI mice livers (n = 4, B); KEGG signaling pathway analysis reveal inflammation related functional terms enriched in HIRI mice at ZT0 and ZT12 (C). (D–F) Mice underwent hepatic I/R surgery at ZT0 after injected with 150 μL clodronate liposomes via the tail vein. Serum transaminase levels and hepatic MPO activity in HIRI mice with macrophages depletion at ZT0 (n = 7, D), the mRNA levels of key cytokines and chemokine in whole liver (n = 7, E), and HE staining and Suzuki scores (n = 7, F) were shown. Scale bar: 50 μm. Data were expressed as mean ± SEM. ∗P < 0.05.