Abstract
Background
Co-infections of the porcine reproductive and respiratory syndrome virus (PRRSV) and the Haemophilus parasuis (HPS) are severe in Chinese pigs, but the immune response genes against co-infected with 2 pathogens in the lungs have not been reported.
Objectives
To understand the effect of PRRSV and/or HPS infection on the genes expression associated with lung immune function.
Methods
The expression of the immune-related genes was analyzed using RNA-sequencing and bioinformatics. Differentially expressed genes (DEGs) were detected and identified by quantitative real-time polymerase chain reaction (qRT-PCR), immunohistochemistry (IHC) and western blotting assays.
Results
All experimental pigs showed clinical symptoms and lung lesions. RNA-seq analysis showed that 922 DEGs in co-challenged pigs were more than in the HPS group (709 DEGs) and the PRRSV group (676 DEGs). Eleven DEGs validated by qRT-PCR were consistent with the RNA sequencing results. Eleven common Kyoto Encyclopedia of Genes and Genomes pathways related to infection and immune were found in single-infected and co-challenged pigs, including autophagy, cytokine-cytokine receptor interaction, and antigen processing and presentation, involving different DEGs. A model of immune response to infection with PRRSV and HPS was predicted among the DEGs in the co-challenged pigs. Dual oxidase 1 (DUOX1) and interleukin-21 (IL21) were detected by IHC and western blot and showed significant differences between the co-challenged pigs and the controls.
Conclusions
These findings elucidated the transcriptome changes in the lungs after PRRSV and/or HPS infections, providing ideas for further study to inhibit ROS production and promote pulmonary fibrosis caused by co-challenging with PRRSV and HPS.
Keywords: Porcine reproductive and respiratory syndrome virus, Haemophilus parasuis, co-infection, immune response, RNA sequencing
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS), caused by the PRRS virus (PRRSV), is one of the most significant and economically important infectious diseases affecting swine worldwide, and the lung is its primary target organ [1]. Additionally, PRRSV is considered one of the main etiological agents in the Porcine Respiratory Disease Complex (PRDC), and the PRDC is a multifactorial respiratory disease complex that predisposes pigs to infection by bacteria such as Mycoplasma hyopneumoniae [2], Haemophilus parasuis (HPS) [3], Streptococcus suis [4], Actinobacillus pleuropneumoniae [5], and Salmonella spp. [6]. HPS is an important conditional pathogen of swine. Its non-virulent strains are common in the upper respiratory tract of pigs, but virulent strains are the causative agent of Glässer's disease, which is involved in the development of PRDC [7].
PRRSV and HPS are resulting in combined infections that exacerbate clinical disease [8]. Single infections with PRRSV or HPS have been studied by comparison of viral or bacterial loads [9], pathogenesis [10,11], clinical syndromes [12], cytokine production [13], and transcriptome analysis [14,15] in pigs or cells. Simultaneous infection with PRRSV and HPS increases inflammatory and immune responses and reduces the production of reactive oxygen species (ROS), which increases the chance of pathogens surviving the host immune response [16,17]. The detailed mechanism and functional genes involved in combined infections have not been studied. Lungs are very important as the first line of defense against invading PRRSV and HPS [18], yet few studies on the response against concurrent PRRSV and HPS infection in pigs at the gene-expression level. Research at the transcriptome level may assist in diagnosing pigs simultaneously infected with PRRSV and HPS and improving understanding of the interactions between pathogens and hosts.
High-throughput sequencing techniques are characterized by high sensitivity, genome-wide coverage, and unbiased quantification of gene transcription [19]. RNA sequencing is a powerful and efficient method to analyze the transcriptome of pigs and to identify immune response genes. This study established experimental models, using PRRSV genotype 2 (Guizhou isolated strain) and HPS serovar 4 (Guizhou isolated strain). All experimental pigs were assessed by clinical symptoms and lung lesions. RNA-sequencing was applied to investigate the gene expression differences in lungs from indigenous Chinese Kele pigs infected with PRRSV or HPS alone or with both pathogens. Filtered differentially expressed genes (DEGs) were validated by quantitative real-time polymerase chain reaction (qRT-PCR), the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs were analyzed. Two selected genes were detected by immunohistochemistry (IHC) and western blotting assays. Our findings provide valuable data for identifying immune response genes, to explain the mechanism of acute inflammatory and immune responses in PRRSV and HPS co-challenged pigs.
MATERIALS AND METHODS
Ethics statement
All animal procedures were conducted following the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Animal Ethics and Research Committee of Guizhou University (Approval No. EAE-GZU-2020-P008). Pentobarbital sodium salt (Sigma, USA) was used to relieve the pain of experimental animals.
Virus and bacterium
The PRRSV GZBJ12 was obtained from the preventive veterinary laboratory at Guizhou University. The GZBJ12 strain with 30aa deletion in nonstructural protein 2 was isolated from a pig herd in the Guizhou province of China in 2017 and belonged to the genotype 2 (North American strain). The titer of the GZBJ12 strain was 105.675 TCID50/mL.
The HPS4 GZ strain was isolated from the Guizhou province of China in 2019 and belonged to serotype 4 (moderately pathogenic strain) confirmed by genotyping test [20]. The concentration of GZ strain inoculum was 1.66 × 109 CFU/mL.
Animal artificial challenge and sample collection
Thirteen 5-week-old Kele piglets from the same litter in a pig farm of Guizhou province were selected for this study. All piglets without PRRSV, HPS, porcine circovirus type 2 (PCV2), or CSFV antigens and antibodies in their blood were identified by PCR and enzyme-linked immunosorbent assay (PRRSV and HPS: Kete Biological Technology Co., Ltd., China; PCV2 and CSFV: Combined Biological Technology Co., Ltd., China). The piglets were randomly divided into 4 groups, housed separately in different isolation rooms, and inoculated pathogens according to a previously described method [21] (Table 1). Piglets were allowed to eat and drink freely. All animals were no secondary infection of other pathogens and euthanized after 10 d post-infection (dpi). Part of the lung tissue of each pig was collected and stored in formaldehyde solution for tissue pathology. Another part of the lung tissue of each pig was collected, homogenized and mixed with Trizol (TaKaRa, Japan) for total RNA and stored at −80°C.
Table 1. The inoculation method of different groups.
| Group | Serial number | Sex | Weight (kg) | Inoculation method |
|---|---|---|---|---|
| PRRSV-HPS | 1 | Male | 21.9 | PRRSV BJ12: IM and IN at a dose of 6.7 × 103.675 TCID50/kg |
| 2 | Female | 15.8 | HPS4: IN at a dose of 2 × 109 CFU/kg | |
| 3 | Female | 18.1 | ||
| 4 | Male | 7.1 | ||
| PRRSV | 1 | Female | 10.5 | PRRSV BJ12: IM and IN at a dose of 6.7 × 103.675 TCID50/kg |
| 2 | Male | 18.4 | ||
| 3 | Male | 15.6 | ||
| HPS | 1 | Female | 12.5 | HPS4: IN at a dose of 2 × 109 CFU/kg |
| 2 | Male | 15.9 | ||
| 3 | Female | 13.7 | ||
| Control | 1 | Male | 18.6 | Inoculated with the same amount of RPMI-1640 |
| 2 | Female | 11.9 | ||
| 3 | Male | 13.1 |
PRRSV BJ12: 105.675 TCID50/mL; HPS4: 1.66 × 109 CFU/mL. RPMI-1640: Gibco, USA.
IM, intramuscular inoculation; IN, intranasal inoculation; PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis.
Clinical symptoms observation and lung lesions evaluation
Rectal temperature and clinical symptoms were recorded daily. Bodyweight was measured on the initial and final days of the experiment. Lung lesions were observed and scored according to 3 criteria [12]. Alveolar epithelial cell necrosis: none (0 point), < 30% (1 point), 30%–60% (2 point), 60%–90% (3 point), > 90% (4 point). Inflammatory cell infiltration: none (0 point), < 30% (1 point), 30%–60% (2 point), 60%–90% (3 point), > 90% (4 point). Thickening of alveolar wall: 1 layer (0 point), 1–2 layers (1 point), 3–4 layers (2 points), 5–6 layers (3 points), > 6 layers (4 points). Each criterion was independently considered and differences between the 4 groups were assessed by Kruskal-Wallis rank sum test and Dunn's test for multiple comparisons. The p < 0.05 was considered significant.
Pathogens load determined by absolute qRT-PCR assay
Total RNA and DNA of each lung were extracted by RNAiso Plus (TaKaRa) and DNAiso Reagent (TaKaRa) for determining PRRSV and HPS loads, respectively. Total RNA was reverse transcribed into cDNA with a PrimeScript RT Master Mix Kit (Perfect Real Time) (TaKaRa). The primers of absolute qRT-PCR were special to the PRRSV N and HPS infB genes [22] as described in Supplementary Table 1. The pUC57 plasmids carrying the PRRSV N and HPS infB genes were proportionally diluted into different concentrations to draw standard curves, respectively. The cDNA or DNA was used for qRT-PCR with a TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa), following the instruction manual. Reactions were performed on an Eppendorf Realplex Sequence Detection System (Eppendorf AG, Germany). Three biological replicate wells of reactions (25 µL) contained 12.5 µL TB Green Premix Ex Taq II (2×), 2 µL of 50 ng/µL cDNA, 1 µL 10 µM of each primer, and 8.5 µL ddH2O. The qRT-PCR program was 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec, 60°C for 30 sec, 72°C for 20 sec, and then we inserted a melt curve stage. The pathogen load of each sample was calculated by the standard curve equation.
RNA preparation and sequencing
RNA degradation and contamination were monitored on 1% agarose gels. RNA purity, concentration, and integrity were measured by a NanoPhotometer spectrophotometer (IMPLEN, USA), Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, USA) and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, USA), respectively. Qualified sample RNA quality met the following conditions: (1) concentration of RNA ≥ 750 ng/μL, (2) total of RNA ≥ 20 μg, (3) optical density (OD)260/280 = 1.8–2.2, (4) RNA integrity number ≥ 8. Following manufacturer recommendations, the total RNA of each swine lung was sequenced with a NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA). The library preparation of each pig (13 pigs in total) was sequenced on an Illumina Hiseq 2500 platform using 125 bp paired-end sequencing reagent kit (Illumina, USA).
RNA-seq data analysis and DEGs identification
Clean reads were obtained by removing reads containing adapter or poly-N and low-quality reads from raw reads; Q20 and Q30 of the clean data were calculated. Clean reads were aligned against ENSEMBL Suscrofa10.2.72 with HISAT2 [23] v2.0.4 running with ‘--rna-strandness RF’; all other parameters were set as default. StringTie [24] v1.3.3 was applied to assemble and calculate Fragments Per Kilobase of exon per Million (FPKMs) of coding transcripts in each sample. FPKM means fragments per kilo-base of exon per million fragments mapped and is calculated based on the length of the fragments and reads count mapped to this fragment. If genes FPKM quality status was not “OK” (FPKM < 0.01) or the FPKM in all samples was zero, it was removed. DEseq2 [25] provided differential expression statistics of the infection and control groups. The Benjamini & Hochberg method (p-adj) was used to adjust the p value. Transcripts with a p-adj < 0.05 and log2|Fold Change| (log2|FC|) ≥ 2 were considered differentially expressed.
Validation of DEGs with qRT-PCR
We used qRT-PCR to identify the DEGs. Total RNA of each lung was extracted, reverse transcribed into cDNA, and detected by qRT-PCR as described above. Eleven DEGs were selected randomly from 3 infected groups after filtering, and the GAPDH gene [26] was used as the internal reference gene. The 2−ΔΔCt method was used to calculate the gene expression fold change. All primers of the selected genes were synthesized by Sangon Biotech (Sangon, China) and showed in Supplementary Table 1.
DEGs functions and interactive network analysis
The DEGs were classified by the GO enrichment categories according to the DAVID database. KOBAS3.0 was used to test the statistical enrichment (p < 0.05) of DEGs in KEGG pathways [27]. Interactive network analysis of DEGs was based on the STRING [28] database, and the gene, which interacts with more than 5 genes, is a hub gene in the network. We constructed networks by extracting the target immune-related gene list from the database using Cytoscape3.7.2 software [29]. The immune response model was constructed using the Pathway build tool 2.0 in the PRRSV-HPS group.
IHC assay
IHC assays were performed as follows: after dehydration, embedding, and slicing of the fixed tissue. Blocked with 3% peroxide-methanol at room temperature for 10 min; rinsed in phosphate-buffered saline (PBS; 3 × 5 min); microwaved 5 minutes in citric acid and finally washed 10 min in PBS. After a 30-min incubation in goat serum (ZLI-9021; Beijing Zhongshan Jinqiao Biotechnology Co., China), sections were incubated with primary antibodies DUOX1 (1:50) (ab230209; Abcam, UK) and IL21 (1:100) (ab5978; Abcam) overnight at 4°C. After 2 rinses in PBS for 10 min, sections were incubated at 37°C with goat anti-rabbit IgG H&L (horseradish peroxidase [HRP]) (ab6721; Abcam) secondary antibodies for 30 min. Coloration with 3,3-diaminobenzidine (K135925C; Beijing Zhongshan Jinqiao Biotechnology Co.), kept at room temperature in darkness for 2 min, finishing coloration with the distilled water. Finally, hematoxylin stained; dehydration, clearing, and mounting with neutral gums.
The stained IHC images were analyzed by Image-Pro software (Media Cybernetics, USA). The intensity of OD measurement and mean densitometric (MD) value calculation details were described in Wang et al. [30]. The results are shown as mean ± SD of 3 views for each sample. Significant differences were assessed by the Wilcoxon rank-sum test, with p < 0.05 considered a significant difference.
Western blot assay
Western blot assays were performed as previously described [15]. The protein was collected from each lung suspended with RIPA lysis buffer (Beyotime, China) from the control and PRRSV-HPS groups. The protein concentration was measured with a BCA protein assay kit (Beyotime). The protein was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel electrophoresis and transferred to polyvinylidene diuoride membranes. After blocking, the membranes were incubated in the primary antibodies IL21 (ab5978; Abcam), DUOX1 (NBP2-16232; Novusbio, USA), GAPDH (GXP199639; GenXspan, USA) and in secondary antibodies IgG H&L (HRP) (ab6721; Abcam). The resulting signals were visualized by an ECL detection kit (KF001; Affinity, USA). Quantitative differences in western blots were analyzed by densitometry analysis using Image J software (National Institutes of Health, USA) [31]. Significant differences were assessed by the Wilcoxon rank-sum test, with p < 0.05 considered a significant difference.
RESULTS
Clinical symptoms and pathology changes
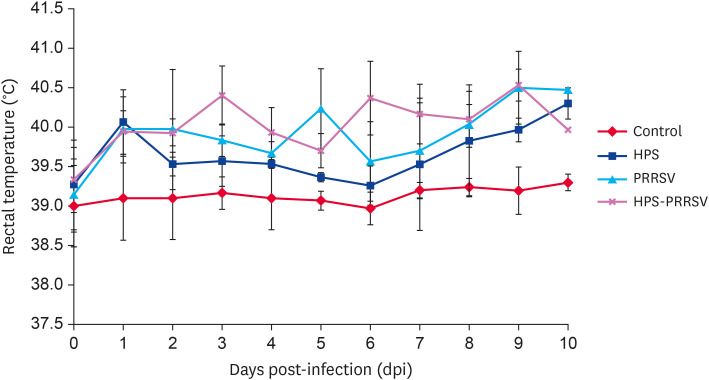
As shown in Fig. 1, the rectal temperature of pigs in the control group ranged from 39.0°C to 39.3°C, with normal average daily gain (0.47 ± 0.01 kg), physical status and behavior. The rectal temperature of pigs in the PRRSV-HPS, PRRSV and HPS groups started to rise at 1st dpi; it was consistently higher than 40°C after 6, 8 and 9 d of infection, respectively. The feed intake of pigs in the PRRSV-HPS, PRRSV and HPS groups decreased, and the average daily weight gain were 0.29 ± 0.18 kg, 0.40 ± 0.12 kg and 0.38 ± 0.07 kg, respectively, which was significantly lower than that of the control group (p < 0.05). In addition, pigs in the 3 infected groups showed different degrees of depression, lying down behavior, increased breathing rate and even abdominal breathing.
Fig. 1. The rectal temperature change in the different groups.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis.
As pigs showed poor physical status and prognosis after consistent fever, all the pigs were slaughtered at 10 dpi to observe lung histopathological changes. Pigs that were not inoculated with pathogens had no microscopic lesions, whereas microscopic lesions of pig lungs in the 3 infected groups showed varying severity of alveolar epithelial cells necrosis, marked thickening of alveolar wall, inflammatory cell infiltration and fibrous hyperplasia (Supplementary Fig. 1). Lung lesion was evaluated by scoring the inflammatory cell infiltration, alveolar wall thickening and alveolar epithelial cell necrosis as described in Table 2. When compared to the control group, pigs in the 3 infected groups had significantly more alveolar epithelial cell necrosis (p < 0.05); pigs in the PRRSV and PRRSV-HPS groups had significantly more inflammatory cell infiltration (p < 0.05) and alveolar wall thickening (p < 0.05). There were no significant differences in lung lesions among the 3 challenge groups.
Table 2. Lung lesion scores of different groups.
| Group | Alveolar epithelial cell necrosis | Inflammatory cell infiltration | Thickness of alveolar wall |
|---|---|---|---|
| Control | 0 ± 0 | 0.33 ± 0.58 | 0.33 ± 0.58 |
| HPS | 1.33 ± 0.58 | 1.33 ± 0.58 | 1.33 ± 0.58 |
| PRRSV | 1.33 ± 0.58 | 2.00 ± 0 | 2.00 ± 0 |
| PRRSV-HPS | 1.50 ± 1.00 | 2.00 ± 0 | 2.00 ± 0 |
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis.
Pathogens load in lung tissue
The lung pathogens load was determined by absolute qRT-PCR. The mean amount of HPS was 105.02 copies/μL and 103.00 copies/μL in the lungs of PRRSV-HPS and HPS groups. The mean amount of PRRSV was 104.91 copies/μL and 103.91 copies/μL in the lungs of PRRSV-HPS and PRRSV groups. No HPS was detected from tissues of control and PRRSV groups, no PRRSV was detected from tissues of control and HPS groups (Supplementary Fig. 2).
Differential expression profiles in pigs infected with different pathogen combinations
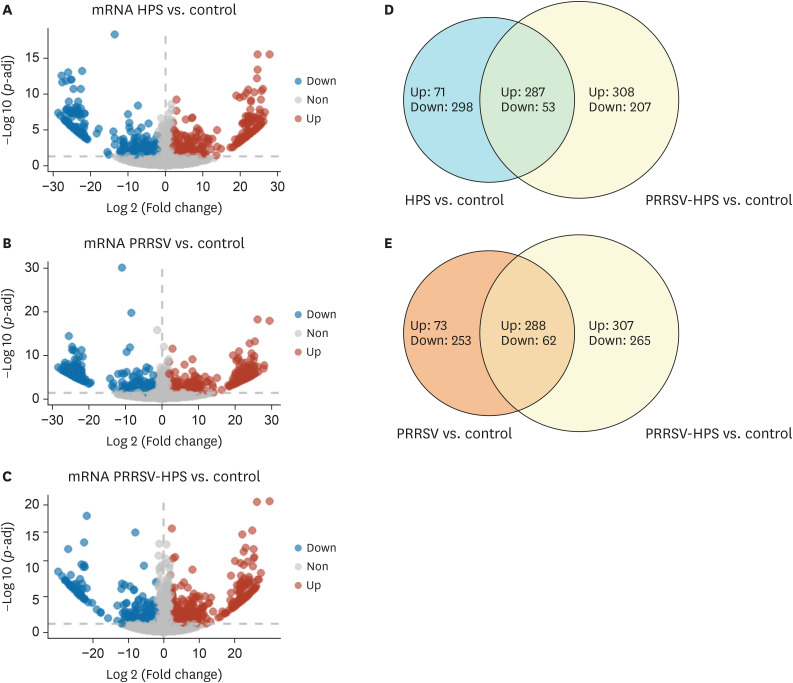
Thirteen cDNA libraries from 4 groups (control, HPS, PRRSV, and PRRSV-HPS groups) were sequenced as described in Supplementary Table 2. DEGs were screened with |log 2(fold change)| ≥ 2 and p-adj < 0.05. A total of 358 genes were up-regulated and 351 genes were down-regulated in pigs with HPS infection. Three hundred sixty-one genes were up-regulated and 315 genes were down-regulated in the PRRSV challenge group. Five hundred ninety-five genes were up-regulated and 327 genes were down-regulated in pigs with PRRSV and HPS infection, indicating that more DEGs existed in co-challenged groups than the 2 single-infected groups (Fig. 2A-C). Among the DEGs in different groups, 340 common DEGs (287 up-regulated genes and 53 down-regulated genes) were present in the HPS and PRRSV-HPS groups, and 582 unique DEGs were present in the concurrent infection group. Three hundred and fifty common DEGs (288 up-regulated and 62 down-regulated) were verified in the PRRSV and PRRSV-HPS groups, and 572 unique DEGs were present in the PRRSV-HPS group (Fig. 2D and E).
Fig. 2. Volcano plots and Venn diagrams of different expression transcripts from HPS, PRRSV, and PRRSV-HPS infected groups. (A-C) Blue indicates significantly ‘Down’, red indicates significantly ‘Up’, and grey means ‘Non’. (D, E) Light blue indicates HPS vs. control DEGs, orange indicates PRRSV vs. control DEGs, yellow indicates PRRSV-HPS vs. control DEGs, and the part of the 2 circles that intersects indicates genes common to co-infection and single infection.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis; Up: up-regulated genes; Down: down-regulated genes; Non: not differentially expressed genes; DEG, differentially expressed gene.
DEG validation by qRT-PCR
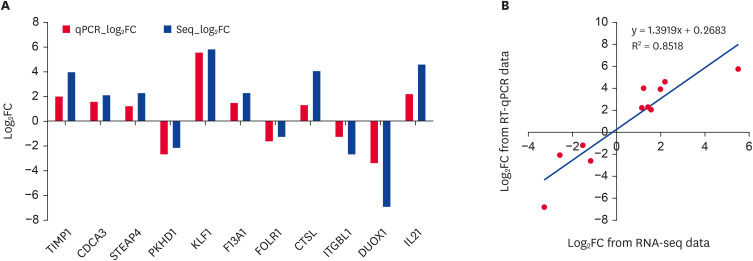
Among the eleven validated DEGs, 7 genes showed a consistent up-regulation trend in virus- or bacteria-challenged pigs, and 4 genes were down-regulated in both the qRT-PCR and RNA-sequencing results (Fig. 3A). Although the extent of fold-change varied between RNA-seq and qRT-PCR, the qRT-PCR confirmed the results of RNA-sequencing, and both methods displayed a strong correlation (R2 = 0.85) (Fig. 3B).
Fig. 3. qRT-PCR identification of randomly selected DEGs and correlation with RNA-seq data. (A) The qRT-PCR identification results of randomly selected DEGs. The X-axis is the name of the genes, and the Y-axis is the log2FC relative expression value; (B) The correlation log2FC from qRT-PCR and RNA-seq assays.
qRT-PCR, quantitative real-time polymerase chain reaction; DEG, differentially expressed gene; FC, Fold Change.
The difference in immune biological processes and KEGG pathways between PRRSV or/and HPS infected pigs
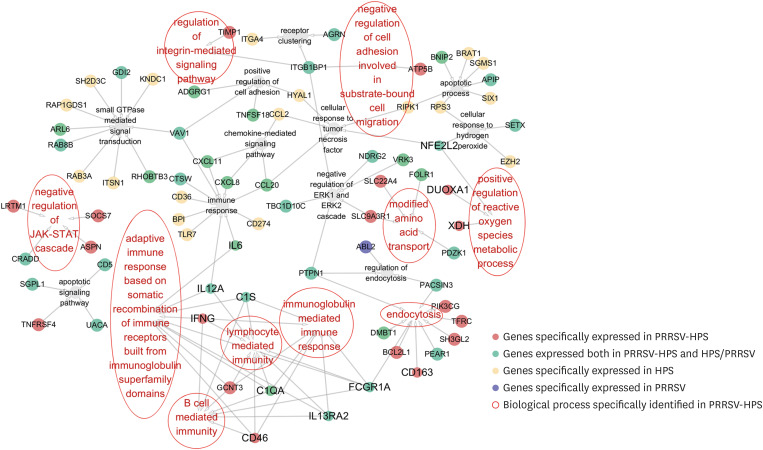
The GO database was used to reveal the differences in GO function and classification of DEGs between PRRSV or HPS alone-infected and co-challenged pigs (Supplementary Table 3A-C). Fourteen immune-related biological processes were annotated in the HPS-infected group, including immune response, receptor cluster, apoptotic process, chemokine mediated signal transduction, and other 8 unique (GO: 0071356, GO: 0070301, GO: 0006955, GO: 0007264, GO: 0045785, GO: 0043113, GO: 0006915, and GO: 0070098) and 2 common biological processes (GO: 0070373, GO: 0006933), as compared with the PRRSV-HPS group, involving 42 genes such as CCL2, CXCL11, CCL20, CXCL8 (IL8), CTSW, CD36, and IL6. The PRRSV-infected group specifically enriched 2 categories, regulation of endocytosis (GO: 0030100) and cellular response to tumor necrosis factor (GO: 0071356), and 3 commons (GO: 0070373, GO: 0006933 and GO: 0097190) biological processes, as compared with the PRRSV-HPS group, including 14 genes such as CD5, CCL1, CCL3L1, and NDRG2. Compared with the 2 single-infected groups, 9 biological processes were specifically enriched in the PRRSV-HPS group, including positive regulation of ROS metabolic process (GO: 2000379), regulation of integrin-mediated signal pathway (GO: 2001044), negative regulation of JAK-STAT cascades (GO: 0046426), and B cell mediated immunity (GO: 0019724), involved xanthine dehydrogenase (XDH), dual oxidase maturation factor 1 (DUOXA)1, SOCS7, CD5, IFNG, and CD46 (Fig. 4).
Fig. 4. Gene ontology (biology process) analysis of DEGs in the lungs of pigs following infection with PRRSV or HPS alone or dual PRRSV-HPS infection.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis; DEG, differentially expressed gene.
The results of KEGG pathways enrichments show that 47 pathways were significantly enriched in PRRSV-HPS challenged pigs, 66 pathways were significantly enriched in PRRSV-challenged pigs, and 92 pathways were significantly identified in the HPS group (Supplementary Table 4A-C). The top 20 pathways in each group were shown in Fig. 5A-C. Among all pathways, 11 common pathways related to infection and immune pathways were found in the 3 groups. The JAK-STAT signaling pathway (including IL13RA2, SOCS7, SOCS2, IL21R, AOX2, IL12A, LEPR, IL21, FHL1, IL11RA, and CREBBP), hematopoietic cell lineage (involving CD5, IL11RA, TFRC, ITGAM, FCGR1A, MME, and CD8B), AMPK signaling pathway (including ACACA, LEPR, PFKM, HMGCR, PFKP, TSC2, STRADA, and PPP2R5B), Wnt signaling pathway (including NFATC3, LRP5, FZD9, BTRC, CTNNB1, PRKACA, CTNND2, APC, and CREBBP), PPAR signaling pathway (FADS2, ACOX2, NR1H3, CYP7A1, GK, and PLIN1), autophagy-animal (involving CTSL, MTMR3, AMBRA1, GABARAP, CFLAR, ATG13, PRKACA, and TSC2), HIF-1 signaling pathway (including TIMP1, HK3, PFKP, PFKM, TFRC, CREBBP, and PGK1), phagosome (including CTSL, FCGR1A, TFRC, ITGAM, SLA-6, COLEC11, and CANX), cytokine-cytokine receptor interaction (including IL13RA2, TNFSF18, CXCL8, IL21R, IL12A, LEPR, IL21, IL11RA, CXCL11, CCL20, and TNFRSF4), antigen processing and presentation (including CD8B, CTSL, CANX, and SLA-6), and adherens junction (including CTNNB1, PTPN1, and CREBBP) were in the PRRSV and HPS concurrent infection pigs. However, the pigs infected with PRRSV or HPS alone had partially different DEGs in the above pathways (Fig. 5D).
Fig. 5. Top 20 KEGG pathway enrichments in the lungs of pigs following infection with PRRSV or HPS alone or dual PRRSV-HPS infection, as well as DEGs of immune-related pathways from co-challenged pigs. (A) HPS challenged group; (B) PRRSV challenged group; (C) PRRSV and HPS co-challenged group; (D) The Circos plot of DEGs list in immune-related KEGG pathways from PRRSV-HPS group.
KEGG, Kyoto Encyclopedia of Genes and Genomes; PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis; DEG, differentially expressed gene.
The immune regulatory network of co-infected pigs was more complicated than pigs infected only with PRRSV or HPS
To explore the similarities and differences of the immune-related DEGs regulatory network between single and concurrent infection pigs, immune-related genes of each infection group were analyzed. The 121 DEGs directly related to immunity, including interleukin family genes, interferon, chemokine family, complements, scavenger receptors family, histocompatibility complex I, and toll-like receptor (Supplementary Table 5) were screened from the different infected groups. Among 80 DEGs in the concurrent-infected group, IL6, IL8, IL21, IL-1β, IFN-γ, CCL11, CCL20, CXCL9, CXCL11, TIMP1, BCL2L1, and C1QA were hub genes. However, TLR7, C1QA, and CD274 played a central role in PRRSV-infected pigs, and IL-1β, IL-8, IL-6, C5, CCL2, CCL20, CCL11, CXCL10, and CXCL11 were hub genes in the HPS-infected immune regulation network (Supplementary Fig. 3).
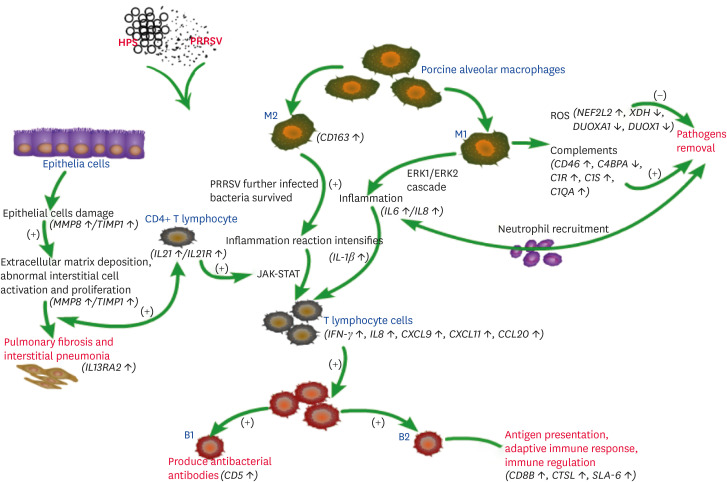
The immune response model was constructed based on the specific biological processes, KEGG pathways, and the immune regulation network enrichments in the co-infected group (Fig. 6). In this model, the host used to defend against PRRSV and HPS invasion was predicted, including directly removing pathogens, promoting lung epithelial-mesenchymal transition, secreting cytokines and chemokines to recruit immune cells and activate downstream pathways.
Fig. 6. The prediction immune response model of PRRSV and HPS co-infection in pigs.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis; ROS, reactive oxygen species.
DEGs related to immune response were identified by IHC and western blot in dual-infected pigs
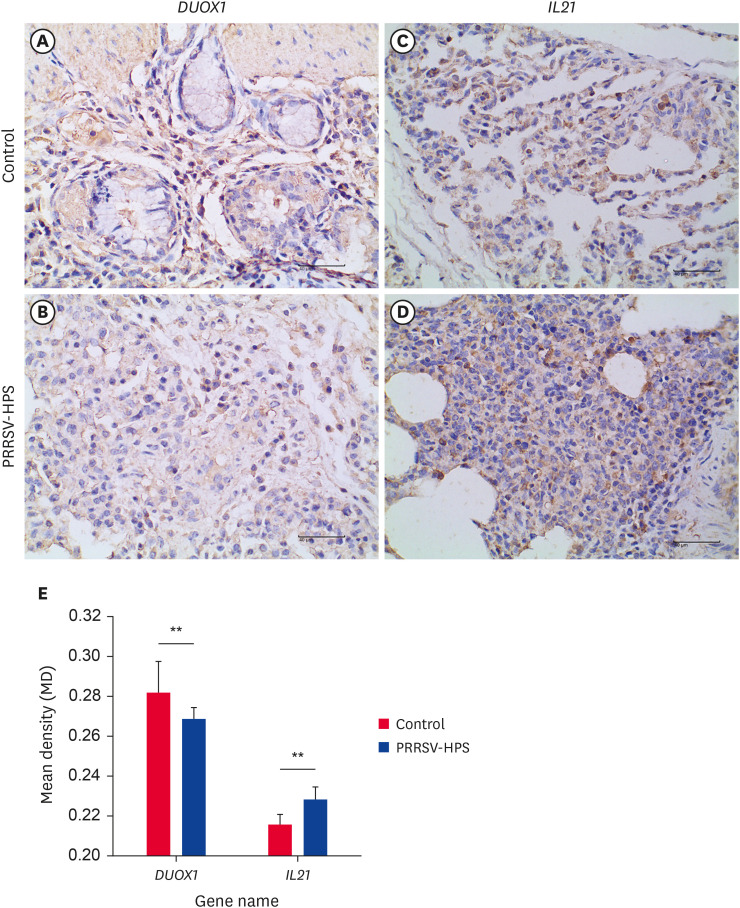
Pigs challenged with PRRSV and HPS, the expression of DUOX1 in the lung tissues was lower than that in the controls (Fig. 7A and B), and the MD of DUOX1 was significantly lower in the co-infected group than in the control group (Fig. 7E). The IL21 in co-infected PRRSV and HPS individuals was higher than in the controls (Fig. 7C and D). Compared with the MD of the control group, the co-infected group displayed a highly significant MD increase (Fig. 7E).
Fig. 7. Immunohistochemical analysis of DUOX1 and IL21 proteins in lungs from the PRRSV-HPS group and the control group. Negative cells are blue, and positive cells are yellow or brown. The DUOX1 in the control group (A) and PRRSV-HPS group (B). The IL21 in the control group (C) and PRRSV-HPS group (D). (E) Mean density of DUOX1 and IL21 proteins in the PRRSV-HPS group and control group.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis.
**p < 0.01 by the Wilcoxon rank-sum test.
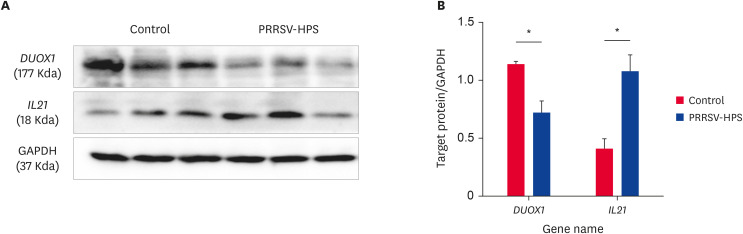
DUOX1 and IL21 were also identified by western blot assay with GAPDH as the housekeeping protein. Pigs simultaneously infected with PRRSV and HPS had a low level of DUOX1 and a high level of IL21 (Fig. 8A); these differences were significant (p < 0.05) compared to the controls (Fig. 8B).
Fig. 8. (A) Western blot analysis of DUOX1 and IL21 proteins in lungs from the PRRSV-HPS and control groups. (A) Detection of DUOX1 and IL21 protein expression by western-blot assay. (B) Relative expression of DUOX1 and IL21 normalized to GAPDH. Every 3 bands are from the same group.
PRRSV, porcine reproductive and respiratory syndrome virus; HPS, Haemophilus parasuis.
*p < 0.05 by the Wilcoxon rank-sum test.
DISCUSSION
According to previous studies, PRRSV infection accelerates HPS infection and lung lesions [17,22,32,33]. Pigs co-infected with HPS serovar 4 (moderate virulence) and swine influenza virus or PCV2 lose weight and exhibit severe lung lesions [21,34]. In this study, pigs infected with single/concurrent PRRSV or HPS showed clinical signs such as fever, reduced daily weight gain, and depression to varying degrees. According to the poor prognosis of the experimental pigs, with the rectal temperature peaking before 9 dpi and significant differences in daily weight gain between the infected and control groups; thus, 10 dpi was determined as a suitable time point to euthanize the pigs for observation. All experimental pigs showed pathological changes. However, pigs infected with HPS showed no enlargement or exudation in the joints, but they had varying degrees of adhesions in the lungs, exudation in the thoracic cavity and pericardium, which may be related to the fact that the HPS strain used was isolated from the lungs. All challenged pigs showed lung lesions. Pigs co-challenged with PRRSV and HPS showed more severe fever, pathological changes and microscopic lung lesions, less daily weight gain and higher pathogens load level than pigs infected with PRRSV or HPS alone. These results suggest that co-infection with PRRSV and HPS exacerbates lung lesions and chronic inflammation, resulting in differential host gene expression.
All experimental pigs were sequenced and DEGs were screened. The qRT-PCR results of eleven differential genes were strongly correlated with the results of RNA-seq (R2 = 0.85), which suggested that the RNA sequencing data met the requirements of subsequent analysis. After analysis and screening, the selected up-regulated gene (IL21) and down-regulated gene (DUOX1) in the PRRSV-HPS group were validated by IHC and western blot assays. These results indicate that PRRSV and HPS invade the porcine respiratory system, regulate gene changes at the transcriptional and protein expression levels to defend against the pathogen infection and further regulate the immune response system.
The lungs of PRRSV-challenged pigs had increased ROS production [35]. However, HPS can inhibit the production of ROS by PAMs to increase bacterial survival [36]. In our study, GO enrichments showed that dual-infection with PRRSV and HPS influences ROS production, consistent with previous studies [16]. Among the ROS genes in our datasets, XDH has been demonstrated to be directly related to ROS levels in mouse ovaries [37]. DUOXA1 is related to the maturation of DUOX1, whose primary function is to catalyze the generation of ROS [38]. Compared to the controls, DUOX1 was down-regulated (log2FC = −6.816) in the concurrent infection group and was not differentially expressed in the single infection group. Compared to the HPS group, DUOX1 was down-regulated (log2FC = −1.821) in the concurrent infection group, while it was not differentially expressed in the concurrently infected group compared to the PRRSV group. So, we infer that the down-regulation of XDH directly inhibited ROS production. The down-regulation of the DUOXA1 gene inhibited ROS production by inhibiting DUOX1 gene expression to prolong pathogen survival in lung tissue, and infection with HPS can indeed down-regulate the expression of DUOX1, and its down-regulation can be accelerated after co-infection with PRRSV. This finding is consistent with Solano et al. [3], but the regulation mechanism requires further study.
PRRSV infection can activate some immune-related KEGG pathways, but the results vary, depending on the animal species [15], the virus strain [39] and the duration of infection [40]. The host takes different strategies to activate immune and inflammatory responses upon HPS infection [41]. In our study, the PRRSV-HPS co-challenged group enrichment immune-related pathways were all in the PRRSV or HPS alone infected groups. Cytokine-cytokine receptor interaction and antigen processing and presentation pathways were enriched in HPS-infected and PRRSV-HPS infected pigs, including up-regulation of IL21 and IL21R. The expression of IL21/IL21R has been detected in human fibrotic lungs [42]. The IL21 (log2FC = 4.564)/IL21R (log2FC = 2.260) genes were significantly up-regulated in the PRRSV-HPS group compared to the controls, while they were not differentially expressed in the concurrently infected group compared to the single infected groups. The specific up-regulation of IL21/IL21R indicates that these gene pairs participate in the process of pulmonary fibrosis caused by concurrent infection. In addition, the up-regulation of matrix metalloproteinases (MMPs) and metalloproteinase tissue inhibitors (TIMPs) is caused by human pulmonary fibrosis [43]. MMP8 (log2FC = 22.825) was upregulated with TIMP1 up-regulation (log2FC = 3.98) in the concurrent infection group. The discovery of MMP8/TIMP1 and IL21/IL21R in respiratory diseases caused by a concurrent porcine virus and bacteria infection provides new ideas for clinical treatment and targeted drug research.
Among the hub genes of the concurrent infection host immune defense model, IL8 is mainly regulated by NF-κB and is associated with PRRSV clearance and lung injury, and it is considered as a candidate gene for antiviral and bacteriostasis [44], which is up-regulated to inhibit PRRSV and HPS infection. IFN-γ plays an antiviral role in coping with PRRSV infection, and compared with susceptible pigs, the up-regulated multiples of local Chinese pigs resistant to PRRSV infection were more obvious [15]. In this study, IFN-γ was up-regulated in the concurrent infection host, and it may have played an anti-infection role after pigs were infected with PRRSV and HPS simultaneously. It is worth noting that CD163, the membrane receptor protein of the PRRSV invasion host, was not up-regulated in the PRRSV single infection pigs, but up-regulated in the PRRSV-HPS concurrent infection group (log2FC = 2.085). However, Martínez-Martínez et al. [45] found that a high expression of CD163 molecule in the lungs of piglets infected with HPS may contribute to HPS clearance. PRRSV-HPS concurrent infection has a superposition effect on CD163, and it is whether promoting PRRSV infection or cleaning HPS is worth in-depth study.
In summary, this study investigated the transcriptome profiles and identified immune response genes in lungs from controls and pigs challenged with PRRSV or/and HPS. In the co-infected group, an immune response model was constructed, and IHC and western blotting assays detected the expression of DUOX1 and IL21. These findings provide ideas for a further study to inhibit ROS production and promote pulmonary fibrosis caused by co-challenged with PRRSV and HPS.
ACKNOWLEDGEMENTS
We thank Dr. Chang Liu (Guizhou University of Traditional Chinese Medicine) for providing guidance on data analysis.
Footnotes
Funding: This study was supported by the Youth Fund of Guizhou Academy of Agricultural Science ([2018]92), the State Key Laboratory of Veterinary Etiological Biology (Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences) (SKLVEB2019KFKT015), and the Guizhou Science and Technology Department (QKHFQ [2018]4007 and QKHZC [2018]2280).
Conflict of Interest: The authors declare no conflicts of interest.
Data Availability Statement: The RNA sequencing data has been uploaded in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA). The accession number is PRJNA661122 (https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA661122).
- Conceptualization: Shi K, Zhang J.
- Investigation: Wang J, Zhang X, Zhao C, Du C, Zhang Y.
- Validation: Wang J, Zhang J.
- Writing - original draft: Zhang J.
- Writing - review & editing: Zhou S, Shi K, Tan Y.
SUPPLEMENTARY MATERIALS
Primers of qRT-PCR pathogen load and for the validation of selected genes
Summary of RNA sequencing data from all pigs
GO enrichments in different groups. A. GO enrichments in the HPS group. B. GO enrichments in the PRRSV group. C. GO enrichments in the PRRSV-HPS group
KEGG pathways with p value < 0.05 in different groups. A. KEGG pathways with p value < 0.05 in the HPS group. B. KEGG pathways with p value < 0.05 in the PRRSV group. C. KEGG pathways with p value < 0.05 in the HPS group
The DEGs associated with immune responses in different groups
Lung tissue lesions in pigs following infection with PRRSV or HPS alone or dual PRRSV-HPS infection using H&E staining. Sections of uninfected control lungs show the normal and clear layers of lung structures (A-B). Lung sections of HPS-infected pigs (C-D). (E-F) shows the lung sections of PRRSV-challenged pigs. Lung sections of PRRSV-HPS co-challenged pigs (G-H). Arrows indicate the histological lesions.
Quantitative analysis of PRRSV and HPS load in lungs via real-time PCR.
The immune-related proteins regulation network. The immune-related proteins regulation network in pigs following infection with HPS (A) or PRRSV (B) alone or dual PRRSV-HPS (C) infection. Yellow circles indicate hub genes for response pathogens challenge.
References
- 1.Shi C, Liu Y, Ding Y, Zhang Y, Zhang J. PRRSV receptors and their roles in virus infection. Arch Microbiol. 2015;197(4):503–512. doi: 10.1007/s00203-015-1088-1. [DOI] [PubMed] [Google Scholar]
- 2.Thanawongnuwech R, Thacker B, Halbur P, Thacker EL. Increased production of proinflammatory cytokines following infection with porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae . Clin Diagn Lab Immunol. 2004;11(5):901–908. doi: 10.1128/CDLI.11.5.901-908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solano GI, Bautista E, Molitor TW, Segales J, Pijoan C. Effect of porcine reproductive and respiratory syndrome virus infection on the clearance of Haemophilus parasuis by porcine alveolar macrophages. Can J Vet Res. 1998;62(4):251–256. [PMC free article] [PubMed] [Google Scholar]
- 4.Thanawongnuwech R, Brown GB, Halbur PG, Roth JA, Royer RL, Thacker BJ. Pathogenesis of porcine reproductive and respiratory syndrome virus-induced increase in susceptibility to Streptococcus suis infection. Vet Pathol. 2000;37(2):143–152. doi: 10.1354/vp.37-2-143. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez Reyes Y, Provost C, Traesel CK, Jacques M, Gagnon CA. Actinobacillus pleuropneumoniae culture supernatant antiviral effect against porcine reproductive and respiratory syndrome virus occurs prior to the viral genome replication and transcription through actin depolymerization. J Med Microbiol. 2018;67(2):249–264. doi: 10.1099/jmm.0.000659. [DOI] [PubMed] [Google Scholar]
- 6.Powell LF, Cheney TE, Williamson S, Guy E, Smith RP, Davies RH. A prevalence study of Salmonella spp., Yersinia spp., Toxoplasma gondii and porcine reproductive and respiratory syndrome virus in UK pigs at slaughter. Epidemiol Infect. 2016;144(7):1538–1549. doi: 10.1017/S0950268815002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Zhang C, Aragon V, Zhou X, Zou M, Wu C, et al. Investigation of Haemophilus parasuis from healthy pigs in China. Vet Microbiol. 2019;231:40–44. doi: 10.1016/j.vetmic.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Z, Qin Y, Lai Z, Peng L, Cai X, Wang L, et al. Microbial ecology of swine farms and PRRS vaccine vaccination strategies. Vet Microbiol. 2012;155(2-4):247–256. doi: 10.1016/j.vetmic.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 9.Macedo N, Rovira A, Oliveira S, Holtcamp A, Torremorell M. Effect of enrofloxacin in the carrier stage of Haemophilus parasuis in naturally colonized pigs. Can J Vet Res. 2014;78(1):17–22. [PMC free article] [PubMed] [Google Scholar]
- 10.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, et al. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. 2016;4(1):129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Tang C, Liao M, Yue H. Update on the pathogenesis of Haemophilus parasuis infection and virulence factors. Vet Microbiol. 2014;168(1):1–7. doi: 10.1016/j.vetmic.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Liang W, Li Z, Wang P, Fan P, Zhang Y, Zhang Q, et al. Differences of immune responses between Tongcheng (Chinese local breed) and large white pigs after artificial infection with highly pathogenic porcine reproductive and respiratory syndrome virus. Virus Res. 2016;215:84–93. doi: 10.1016/j.virusres.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 13.de la Fuente AJ, Ferri EF, Tejerina F, Frandoloso R, Martínez SM, Martín CB. Cytokine expression in colostrum-deprived pigs immunized and challenged with Haemophilus parasuis . Res Vet Sci. 2009;87(1):47–52. doi: 10.1016/j.rvsc.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Fu S, Guo J, Li R, Qiu Y, Ye C, Liu Y, et al. Transcriptional profiling of host cell responses to virulent Haemophilus parasuis: new insights into pathogenesis. Int J Mol Sci. 2018;19(5):1320. doi: 10.3390/ijms19051320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang W, Ji L, Zhang Y, Zhen Y, Zhang Q, Xu X, et al. Transcriptome differences in porcine alveolar macrophages from Tongcheng and large white pigs in response to highly pathogenic porcine reproductive and respiratory syndrome virus (PRRSV) infection. Int J Mol Sci. 2017;18(7):1475. doi: 10.3390/ijms18071475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kavanová L, Matiašková K, Levá L, Štěpánová H, Nedbalcová K, Matiašovic J, et al. Concurrent infection with porcine reproductive and respiratory syndrome virus and Haemophilus parasuis in two types of porcine macrophages: apoptosis, production of ROS and formation of multinucleated giant cells. Vet Res (Faisalabad) 2017;48(1):28. doi: 10.1186/s13567-017-0433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Wang S, Li C, Wang C, Liu Y, Wang G, et al. Secondary Haemophilus parasuis infection enhances highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infection-mediated inflammatory responses. Vet Microbiol. 2017;204:35–42. doi: 10.1016/j.vetmic.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10(6):453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia A, Zhou R, Fan H, Yang K, Zhang J, Xu Y, et al. Development of serotype-specific PCR assays for typing of Haemophilus parasuis isolates circulating in Southern China. J Clin Microbiol. 2017;55(11):3249–3257. doi: 10.1128/JCM.00688-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Li W, Wang Y, Gu C, Liu X, Charreyre C, et al. Coinfection with Haemophilus parasuis serovar 4 increases the virulence of porcine circovirus type 2 in piglets. Virol J. 2017;14(1):227. doi: 10.1186/s12985-017-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Wu J, Zhang Y, Guo L, Cong X, Du Y, et al. Concurrent highly pathogenic porcine reproductive and respiratory syndrome virus infection accelerates Haemophilus parasuis infection in conventional pigs. Vet Microbiol. 2012;158(3-4):316–321. doi: 10.1016/j.vetmic.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varet H, Brillet-Guéguen L, Coppée JY, Dillies MA. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One. 2016;11(6):e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feiden S, Wolfrum U, Wegener G, Kamp G. Expression and compartmentalisation of the glycolytic enzymes GAPDH and pyruvate kinase in boar spermatogenesis. Reprod Fertil Dev. 2008;20(6):713–723. doi: 10.1071/rd08004. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(Web Server issue):W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47(1):8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang CJ, Zhou ZG, Holmqvist A, Zhang H, Li Y, Adell G, et al. Survivin expression quantified by Image Pro-Plus compared with visual assessment. Appl Immunohistochem Mol Morphol. 2009;17(6):530–535. doi: 10.1097/PAI.0b013e3181a13bf2. [DOI] [PubMed] [Google Scholar]
- 31.Gallo-Oller G, Ordoñez R, Dotor J. A new background subtraction method for Western blot densitometry band quantification through image analysis software. J Immunol Methods. 2018;457:1–5. doi: 10.1016/j.jim.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Kavanová L, Prodělalová J, Nedbalcová K, Matiašovic J, Volf J, Faldyna M, et al. Immune response of porcine alveolar macrophages to a concurrent infection with porcine reproductive and respiratory syndrome virus and Haemophilus parasuis in vitro . Vet Microbiol. 2015;180(1-2):28–35. doi: 10.1016/j.vetmic.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Kavanová L, Matiašková K, Levá L, Nedbalcová K, Matiašovic J, Faldyna M, et al. Concurrent infection of monocyte-derived macrophages with porcine reproductive and respiratory syndrome virus and Haemophilus parasuis: a role of IFNα in pathogenesis of co-infections. Vet Microbiol. 2018;225:64–71. doi: 10.1016/j.vetmic.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 34.Pomorska-Mól M, Dors A, Kwit K, Czyżewska-Dors E, Pejsak Z. Coinfection modulates inflammatory responses, clinical outcome and pathogen load of H1N1 swine influenza virus and Haemophilus parasuis infections in pigs. BMC Vet Res. 2017;13(1):376. doi: 10.1186/s12917-017-1298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binjawadagi B, Dwivedi V, Manickam C, Torrelles JB, Renukaradhya GJ. Intranasal delivery of an adjuvanted modified live porcine reproductive and respiratory syndrome virus vaccine reduces ROS production. Viral Immunol. 2011;24(6):475–482. doi: 10.1089/vim.2011.0040. [DOI] [PubMed] [Google Scholar]
- 36.Eason MM, Fan X. The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog. 2014;74:50–58. doi: 10.1016/j.micpath.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Liu JC, Li L, Yan HC, Zhang T, Zhang P, Sun ZY, et al. Identification of oxidative stress-related Xdh gene as a di(2-ethylhexyl)phthalate (DEHP) target and the use of melatonin to alleviate the DEHP-induced impairments in newborn mouse ovaries. J Pineal Res. 2019;67(1):e12577. doi: 10.1111/jpi.12577. [DOI] [PubMed] [Google Scholar]
- 38.Konaté MM, Antony S, Doroshow JH. Inhibiting the activity of NADPH oxidase in cancer. Antioxid Redox Signal. 2020;33(6):435–454. doi: 10.1089/ars.2020.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Y, An TQ, Tian ZJ, Wei TC, Jiang YF, Peng JM, et al. The gene expression profile of porcine alveolar macrophages infected with a highly pathogenic porcine reproductive and respiratory syndrome virus indicates overstimulation of the innate immune system by the virus. Arch Virol. 2015;160(3):649–662. doi: 10.1007/s00705-014-2309-7. [DOI] [PubMed] [Google Scholar]
- 40.Zhen Y, Wang F, Liang W, Liu J, Gao G, Wang Y, et al. Identification of differentially expressed non-coding RNA in porcine alveolar macrophages from Tongcheng and large white pigs responded to PRRSV. Sci Rep. 2018;8(1):15621. doi: 10.1038/s41598-018-33891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Liu C, Fang Y, Liu X, Li W, Liu S, et al. Transcription analysis on response of porcine alveolar macrophages to Haemophilus parasuis . BMC Genomics. 2012;13(1):68. doi: 10.1186/1471-2164-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parrish-Novak J, Foster DC, Holly RD, Clegg CH. Interleukin-21 and the IL-21 receptor: novel effectors of NK and T cell responses. J Leukoc Biol. 2002;72(5):856–863. [PubMed] [Google Scholar]
- 43.Odaka C, Tanioka M, Itoh T. Matrix metalloproteinase-9 in macrophages induces thymic neovascularization following thymocyte apoptosis. J Immunol. 2005;174(2):846–853. doi: 10.4049/jimmunol.174.2.846. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Du Y, Wang H, Du L, Feng WH. Porcine reproductive and respiratory syndrome virus (PRRSV) up-regulates IL-8 expression through TAK-1/JNK/AP-1 pathways. Virology. 2017;506:64–72. doi: 10.1016/j.virol.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martínez-Martínez S, Rodríguez-Ferri EF, Frandoloso R, Garrido-Pavón JJ, Zaldívar-López S, Barreiro C, et al. Molecular analysis of lungs from pigs immunized with a mutant transferrin binding protein B-based vaccine and challenged with Haemophilus parasuis . Comp Immunol Microbiol Infect Dis. 2016;48:69–78. doi: 10.1016/j.cimid.2016.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers of qRT-PCR pathogen load and for the validation of selected genes
Summary of RNA sequencing data from all pigs
GO enrichments in different groups. A. GO enrichments in the HPS group. B. GO enrichments in the PRRSV group. C. GO enrichments in the PRRSV-HPS group
KEGG pathways with p value < 0.05 in different groups. A. KEGG pathways with p value < 0.05 in the HPS group. B. KEGG pathways with p value < 0.05 in the PRRSV group. C. KEGG pathways with p value < 0.05 in the HPS group
The DEGs associated with immune responses in different groups
Lung tissue lesions in pigs following infection with PRRSV or HPS alone or dual PRRSV-HPS infection using H&E staining. Sections of uninfected control lungs show the normal and clear layers of lung structures (A-B). Lung sections of HPS-infected pigs (C-D). (E-F) shows the lung sections of PRRSV-challenged pigs. Lung sections of PRRSV-HPS co-challenged pigs (G-H). Arrows indicate the histological lesions.
Quantitative analysis of PRRSV and HPS load in lungs via real-time PCR.
The immune-related proteins regulation network. The immune-related proteins regulation network in pigs following infection with HPS (A) or PRRSV (B) alone or dual PRRSV-HPS (C) infection. Yellow circles indicate hub genes for response pathogens challenge.