Table 3.
The pScore and inDrug values from Badapple data base, and the predicted ADMET properties of JK-KI3.
| Compound | Scaffold Number | Scaffold Structure | pScorea | inDrugb |
|---|---|---|---|---|
| JN-KI3 | Scaf1 | 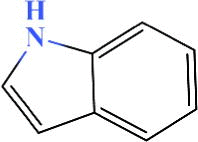 |
370 | True |
| Scaf2 | 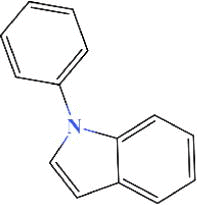 |
186 | True | |
| Scaf3 | 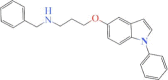 |
None | False | |
| Scaf4 | 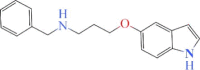 |
None | False |
| ADMET Prediction | |||||
| Propertiesc | Value | Optimal range | Properties | Value | Optimal range |
| CNS | 1 | −2 - +2 | polrz | 51.04 | 13.0–70.0 |
| logPC16 | 16.27 | 4.0–18.0 | logPoct | 23.54 | 8.0–35.0 |
| logPw | 11.51 | 4.0–45.0 | logPo/w | 5.53 | −2.0–6.5 |
| logS | −6.31 | −6.5–0.5 | CIlogS | −6.67 | −6.5–0.5 |
| logHERG | −7.53 | concern below −5 | PCaco | 364.52 | <25 poor,>500 great |
| logBB | −0.37 | −3.0–1.2 | PMDCK | 453.07 | <25 poor,>500 great |
| logKp | −3.08 | −8.0 - −1.0 | logKhsa | 1.05 | −1.5–1.5 |
| PHOA | 92.20 | >80% is high<25% is poor | rtvFG | 0 | 0–2 |
pScore values advisory: <100 (low), means no indication; 100–300 (moderate), means weak indication of promiscuity; >300 (high), means strong indication of promiscuity.
inDrug results: true, means it was found in the drug data base; false, means not found.
properties: CNS, predicted central nervous system activity on a −2 (inactive) to + 2 (active) scale; Polrz, predicted polarizability in cubic angstroms; logPC16, predicted hexadecane/gas partition coefficient; logPoct, predicted octanol/gas partition coefficient; logPw, predicted water/gas partition coefficient; logPo/w, predicted octanol/water partition coefficient; logS, predicted aqueous solubility; CIlogS, conformation-independent predicted aqueous solubility; logHERG, predicted IC50 value for blockage of HERG K+ channels; PCaco, predicted apparent Caco-2 cell permeability in nm/sec; logBB, predicted brain/blood partition coefficient; PMDCK, predicted apparent MDCK cell permeability in nm/sec. logKp, predicted skin permeability; logKhsa, prediction of binding to human serum albumin; PHOA, predicted human oral absorption on 0 to 100% scale; rtvFG, Number of reactive functional groups.
