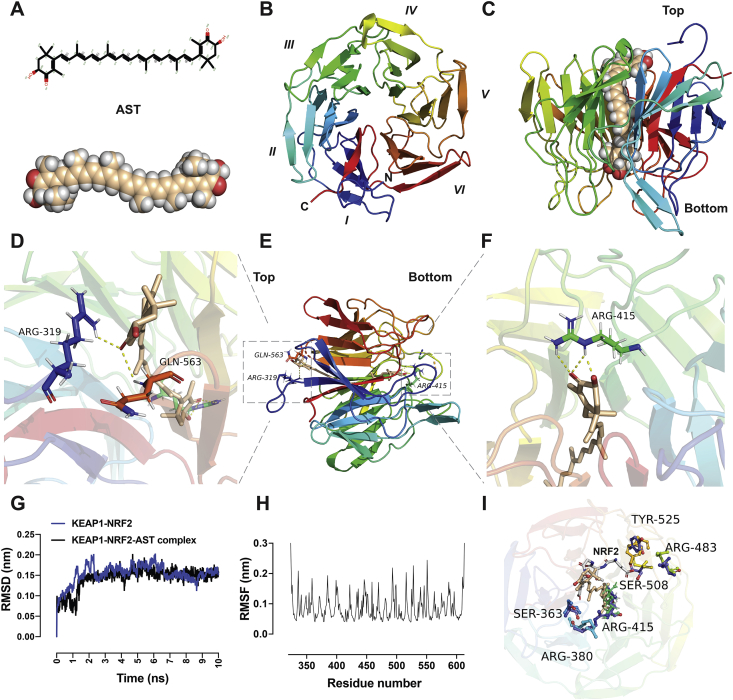
Figure 1.
Molecular docking and dynamic analysis of astaxanthine and KEAP1. (A) Molecular structure of astaxanthine. (B) A cartoon representation of the KEAP1–NRF2 (PDB ID: 6LRZ) β-propeller domain is shown from the top view. The propeller domain contains six blades folded into a pseudo-six-fold symmetry. (C) A cartoon representation of KEAP1 in complex with astaxanthine molecule at the center of the protein from the side view. (D)–(F) A cartoon representation of intermolecular interactions between KEAP1–NRF2 and astaxanthine from different views. Astaxanthine exerts hydrophilic interactions with the binding site residues at the top (D) and bottom (F) sites. The important residues establishing the hydrophilic interactions are Arg 319, Arg 415, and Gln 563. (G) RMSD of the KEAP1–NRF2–AST complex and KEAP1–NRF2 obtained within molecular dynamics simulation for 10 ns. (H) The time-averaged RMSF of KEAP1–NRF2 residues was obtained to analyse the local mobility of the protein after docking. (I) Comparison between the KEAP1–NRF2–AST complex and the KEAP1–NRF2 (PDB ID: 5WFV) reveals that astaxanthine partially overlapped with the ETGE motif of the NRF2 peptide. The NRF2-binding sites reveal significant changes in the orientation of residues including Arg 380, Arg415, Arg 483, Ser 508, and Tyr525 after AST docking to KEAP1–NRF2. The figures are generated using PyMOL. AST, astaxanthine; PDB, protein data bank; RMSD, root mean square deviation; RMSF, root mean square fluctuations.