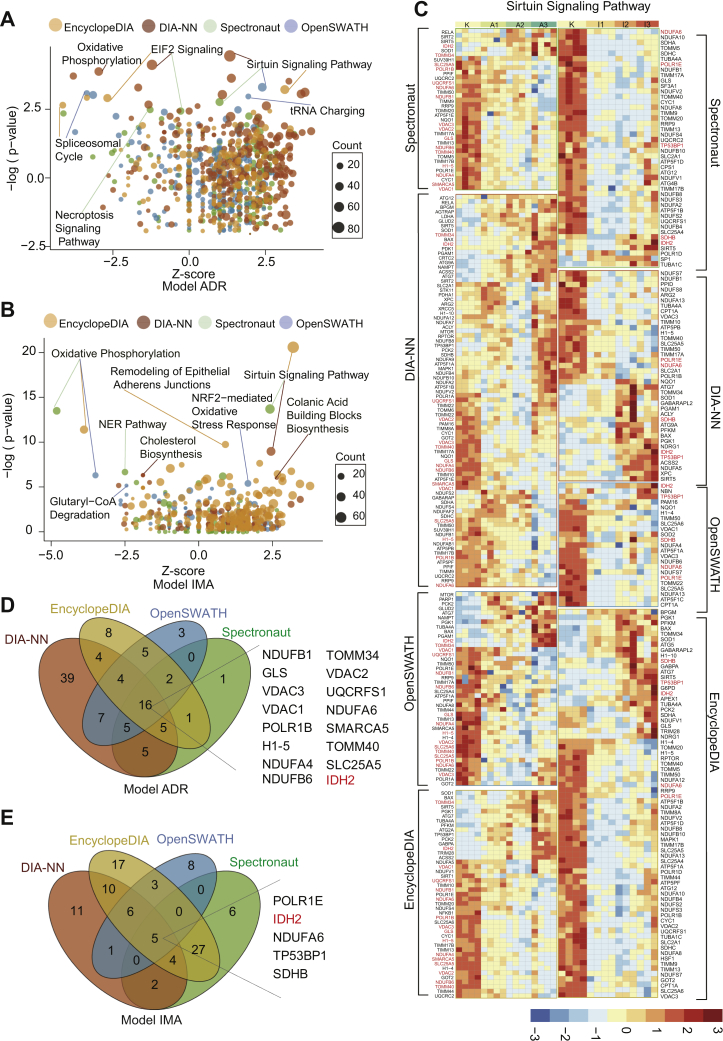
Fig. 5.
Enriched proteins that exhibit continuous changes as identified by IPA in ADR-resistant and IMA-resistant cells.A, in the ADR-resistant cells, the three most significantly changed pathways were enriched by IPA from four DIA software. B, in the IMA-resistant cells, the three most significantly changed pathways were enriched by IPA from four DIA software. C, the heatmap shows the proteins (adjusted p value < 0.05 calculated by ANOVA) involved in sirtuin signaling pathway from four DIA software expression in the IMA-resistant and ADR-resistant cells, with the overlapped quantified proteins being highlighted in red. Each row indicats a protein, and each column indicats a sample. The protein intensity matrix was normalized by Z-score and colored in the heatmap. D, Venn diagram shows the 16 overlapped proteins involved in the sirtuin signaling pathway in the ADR-resistant cells. E, Venn diagram shows the five overlapped proteins involved in the sirtuin signaling pathway in the IMA-resistant cells. ADR, adriamycin; DIA, data-independent acquisition; IMA, imatinib; IPA, ingenuity pathway analysis.