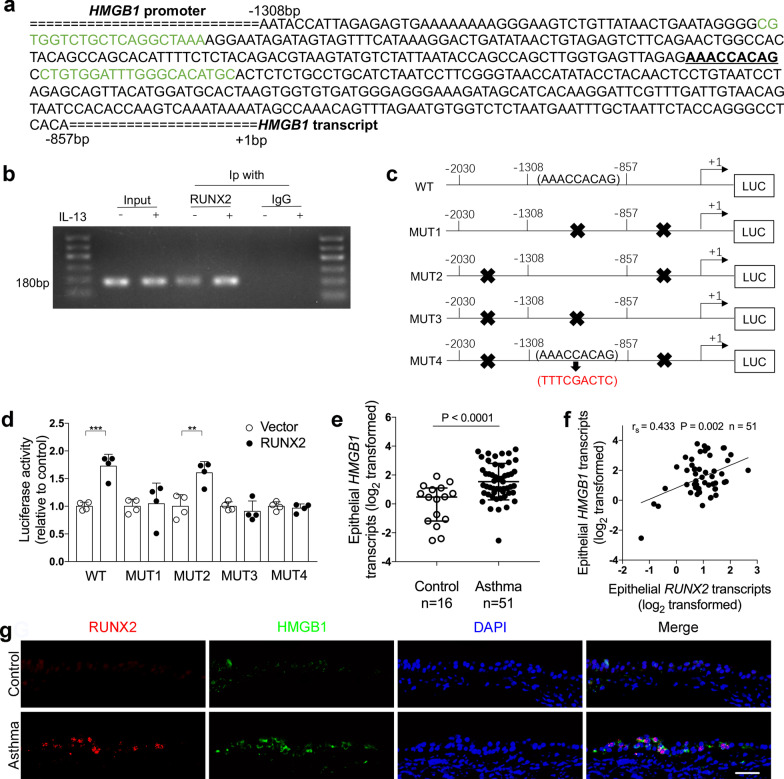
Fig. 4.
RUNX2 binds to the promoter of HMGB1. a The promoter region of HMGB1 has a putative binding site for RUNX2. Sequence scheme of HMGB1 promoter region with the putative RUNX2 binding element underlined (AAACCACAG). Sequences marked in green are primers for ChIP-PCR assay in panel B. b ChIP-PCR assays to amplify the 180-bp region of HMGB1 promoter were performed to show direct binding of RUNX2 to HMGB1 promoter in BEAS-2B cells. c Schematic presentation showing the luciferase reporter plasmid containing the wild type (WT), truncated, or mutant HMGB1 promoter (MUT1–MUT4). MUT1 retains − 2030 to − 1308 bp, MUT2 retains − 1308 to − 857 bp, MUT3 retains − 857 to + 1 bp. MUT4 retains − 1308 to − 857 bp and the putative RUNX2 binding element AAACCACAG was replaced with TTTCGACTC. d The luciferase reporter plasmids containing WT and mutant HMGB1 promoter were co-transfected with empty or RUNX2 cDNA expression vector. Luciferase activity was measured by dual-luciferase reporter assay system. The renilla luciferase activity was normalized to firefly luciferase activity (one-way ANOVA followed by Tukey’s multiple comparison test). n = 4 wells per group. Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001. e HMGB1 mRNA levels in bronchial brushings from asthma patients (n = 51) and controls (n = 16) were determined by quantitative PCR assays. The mRNA levels were expressed as log2 transformed and relative to the median value of controls (two‐tailed Mann–Whitney test). f Correlation assays between epithelial transcript levels of RUNX2 and HMGB1 in asthma patients (n = 51). Correlation assays were performed using Spearman's rank‐order correlation. g Representative images of RUNX2 (red) and HMGB1 (green) immunofluorescence staining in bronchial biopsies from controls and asthma patients. Nuclei were stained with DAPI (blue). Scale bar, 50 μm