Figure 5. PPIL1 P95 is positioned in the enzymatic pocket of PRP17 in the activated spliceosome.
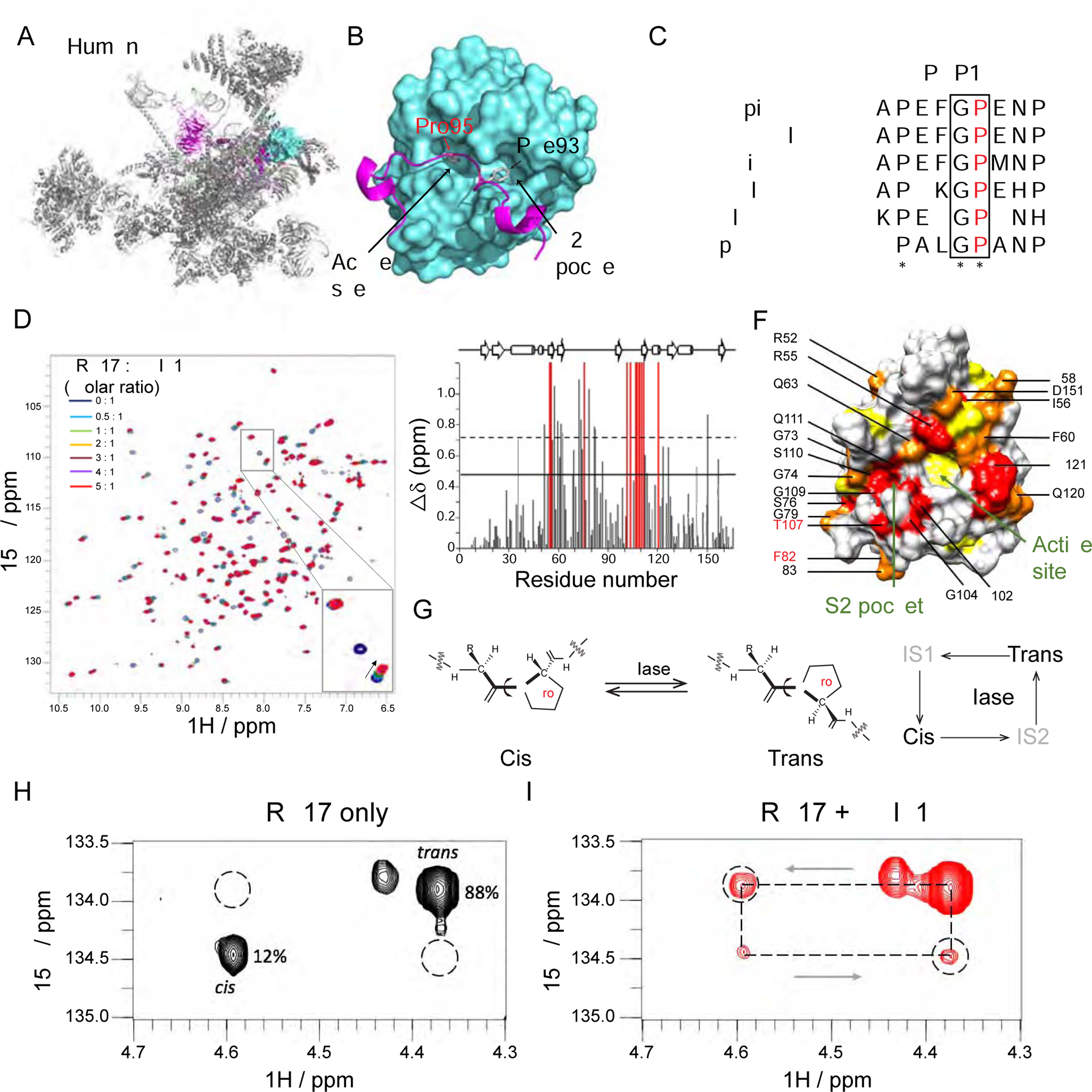
(A and B) Cryo-EM structure of human spliceosome C* complex (PDB: 5XJC) shows an N-terminal loop of PRP17 (cartoon in purple) bound to PPIL1 (teal) enzymatic surface with Pro95 buried inside the S1 enzymatic pocket.
(C) Protein sequence alignment of PRP17 from 6 species shows an evolutionarily conserved Gly-Pro (G-P) motif in PRP17. * stands for identical residues, : indicates similar residues.
(D) Overlaid 1H-15N HSQC spectra of PPIL1 with PRP17 peptide titrations (0–5 molar equivalents, aa 89–101: FAPEFG[P]ENPFRT). Specific resonance shifts or broadening beyond detection indicates specific binding of PRP17 to PPIL1. Inset: Examples of PPIL1 resonance changes that shift (arrowhead), broaden beyond detection (open arrowhead) or were unchanged (arrow) as a result of PRP17 titration.
(E) Average chemical shift perturbations of PPIL1 residues upon titration with PRP17. Gray: shifted resonances. Red: broadened beyond detection resonances. Solid or dashed lines: shifts >1 or >2 SD above mean, respectively.
(F) Space-filling model of PPIL1 (PDB: 2X7K) showed significantly perturbed residues upon PRP17 peptide binding. Red: residues broadened beyond detection, Orange: residues >2 SD, Yellow: residues >1 SD above mean. Residues affected in patients are labeled in red.
(G) Schematic of cis-trans Xaa-Pro peptide bond isomerization catalyzed by PPIase. IS: Intermediate State.
(H and I) 2D 1H, 15N-H(Cα)N ZZ exchange spectra of PRP1 peptide in the absence (H) or presence (I) of sub-stoichiometric concentrations (1% molar ratio) of PPIL1, with appearance of ‘exchange signals’ (dashed circles), i.e. significant interconversion between cis-trans states.
See also Figures S5 and S6, Table S4, and Movie S1.
