FIGURE 2.
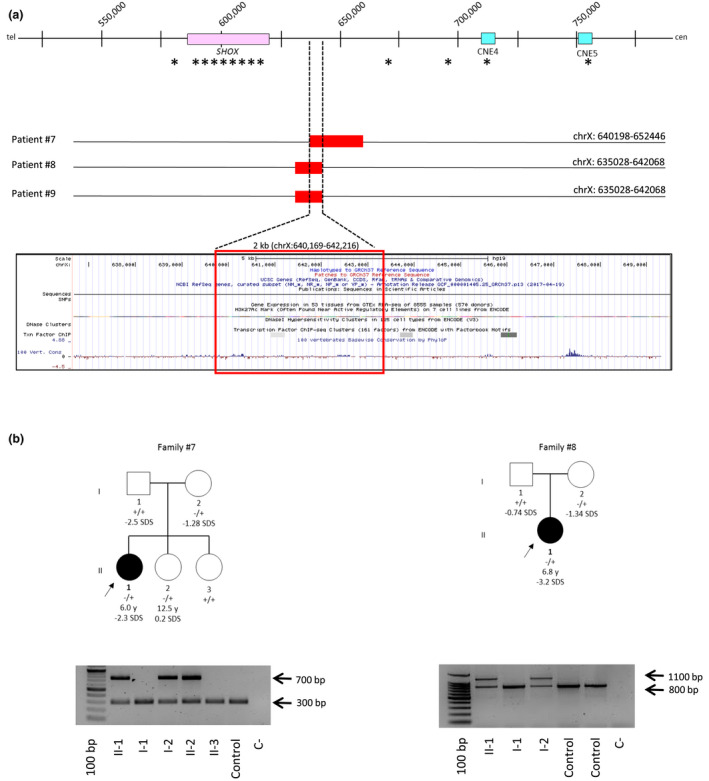
(a) Deletions identified through custom array CGH in patients #7, #8, #9. The asterisks indicate the MLPA probes. The UCSC Genome Browser shows poor evolutionary conservation within the minimal deleted region is indicated. (b) Pedigrees of patients #7 and #8. The parents of patient #9 were not available for the analysis. The proband is indicated by an arrow. The wild‐type and variant alleles are indicated as − and +, respectively. A PCR assay was specifically designed to detect each of the deletions identified through the fine tiling aCGH. The forward and the reverse primers were designed just upstream and downstream the putative 5′ and 3′ breakpoints, respectively; the PCR product is detectable only in deletion carriers (upper band). A further reverse primer was designed within the deleted sequence to amplify wild‐type alleles (lower band)
