Figure 2.
CRISPR-Cas3-based assay for rapid detection of SARS-CoV-2 and influenza virus
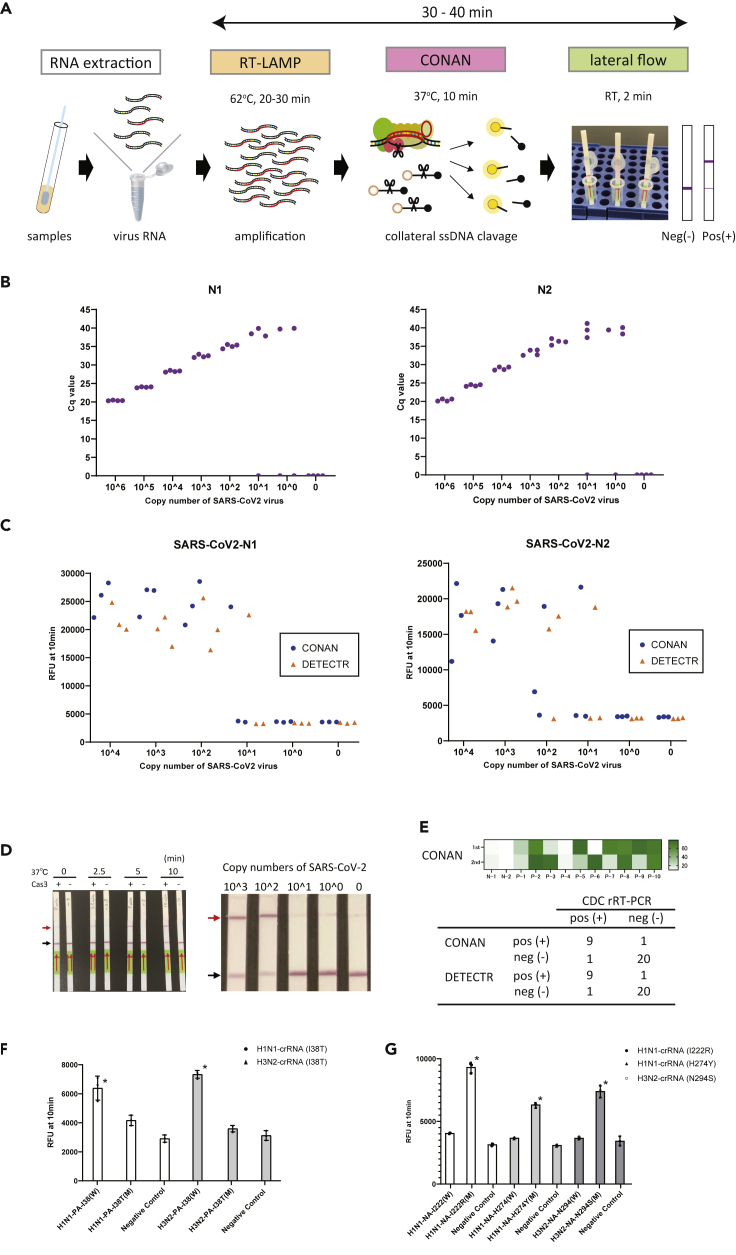
(A) Schematic representation of the CONAN SARS-CoV-2 detection assay including a conventional RNA extraction step, RT-LAMP (62 °C, 20–30 min), CONAN (37 °C, 10 min), and lateral flow (RT, 2 min).
(B) The limit of detection (LoD) of the US CDC’|'s RT-qPCR assay amplification of the N1 and N2 regions of the SARS-CoV-2 N gene. Cq, cycle quantification value.
(C) LoD of CONAN-based and DETECTR-based assays for the N1 and N2 region of SARS-CoV-2. RFU, relative fluorescence unit.
(D) LoD of the CONAN-based lateral flow assay for the N1 region of SARS-CoV-2. CONAN’|'s LoD was 2 min incubation (left) and <102 copies (right). Positive (red arrow) and negative (black arrow) bands for CONAN (see Figure S8).
(E) Comparison of SARS-CoV-2 CONAN and DETECTR assays on 31 clinical samples (10 positive and 21 negative for SARS-CoV-2 by the CDC RT-qPCR assay) (see also Figure S10).
(F) CONAN-based assay for detecting I38T variants in influenza viruses. Means (n = 3) and standard deviations. ∗p < 0.01, one-way ANOVA with post-hoc test.
(G) CONAN-based assay for detecting I222R, H274Y, and N294S variants in influenza viruses. Means (n = 3) and standard deviations. ∗p < 0.01, one-way ANOVA with post-hoc test.