Figure 1. Loss of CHMP7 does not impair MVE biogenesis or ESCRT-mediated endosomal protein sorting.
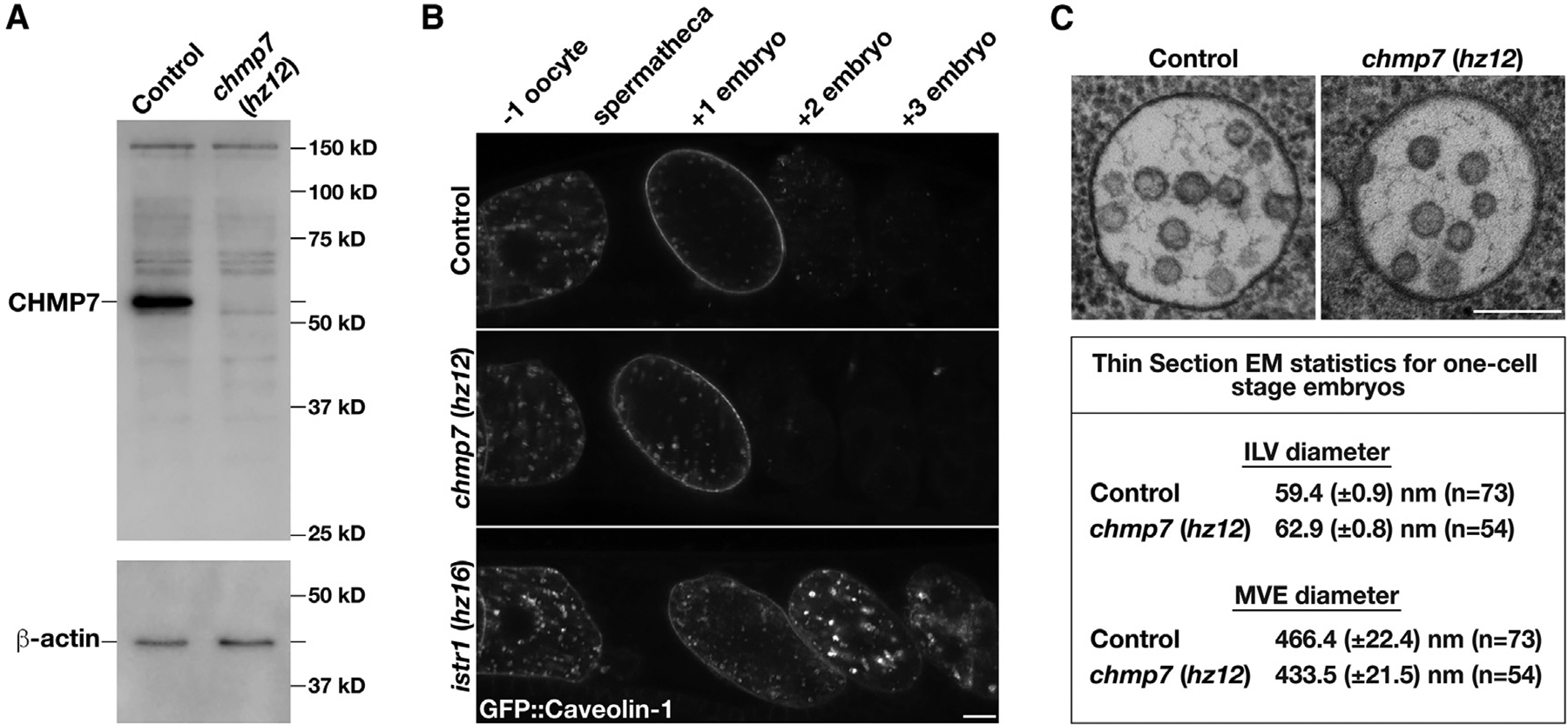
(A) Representative immunoblot of extracts generated from control and chmp7 (hz12) mutant animals (n = 3 each) using antibodies directed against CHMP7 (top) and beta-actin (bottom; load control).
(B) Anesthetized control and CRISPR-Cas9-edited animals harboring deletion mutations in CHMP7 or ISTR1 and expressing a GFP fusion to C. elegans caveolin-1 were imaged using swept-field confocal microscopy. Caveolin-1 is normally degraded prior to embryos reaching the two-cell stage (Frankel et al., 2017). The relative positions of oocytes and embryos are highlighted. Images are representative of at least 10 animals of each genotype. Bar, 10 μm.
(C) High-pressure frozen control and chmp7 (hz12) mutant animals were processed for thin section electron microscopy to visualize MVEs within the one-cell stage embryo. Micrographs are representative of at least 50 MVEs. Bar, 200 nm. A summary of MVE and ILV sizes is also shown.
See also Figure S1.
