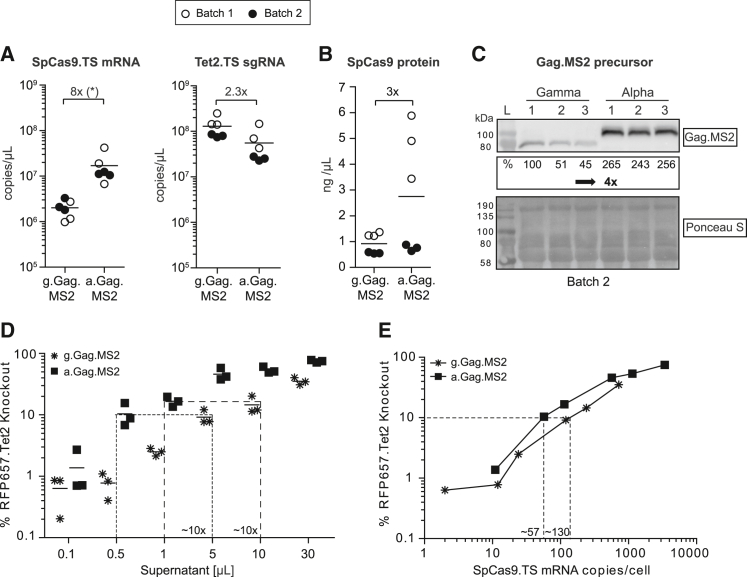
Figure 3.
Characterization of alpha- and gammaretroviral Gag.MS2.CRISPR.Tet2 supernatants
In total, two different batches (white and black filled circles), each with three individually packaged supernatants for a.Gag.MS2 and g.Gag.MS2 CRISPR.Tet2 particles, were generated and characterized for their content of CRISPR-Cas9 components (n = 6). (A) SpCas9.TS mRNA and sgRNA.TS content in a.Gag.MS2- and g.Gag.MS2-based CRISPR.Tet2 supernatants. Total RNA copies per microliter supernatant were calculated using individual plasmid standards. (B) Gag.MS2.CRISPR.Tet2 supernatants contain SpCas9 protein. SpCas9 protein concentrations were assessed by an SpCas9 ELISA. (C) a.Gag.MS2.CRISPR.Tet2 supernatants had a higher Gag.MS2 precursor protein content. Immunoblot analysis of three individually packaged a.Gag.MS2.CRISPR.Tet2 and g.Gag.MS2.CRISPR.Tet2 supernatants is shown (batch 2; black filled circles) (n = 3). g.Gag.MS2 (82 kDa) and a.Gag.MS2 (89 kDa) precursor proteins were detected with an anti-MS2 antibody. Ponceau S staining of the membrane is shown below. (D) g.Gag.MS2.CRISPR.Tet2-mediated RFP657.Tet2 knockout rates can be adjusted by ∼10-fold higher supernatant volumes. Characterized supernatants from batch 2 were used to transduce HT1080 RFP657.Tet2 reporter cells at the depicted volumes (n = 3). (E) Normalization of supernatant volumes showed an ∼2-fold-higher potency for a.Gag.MS2-based CRISPR.Tet2 particles. The in (D) acquired datasets were replotted as % RFP657.Tet2 knockout against applied SpCas9.TS mRNA copies/cell (n = 3; mean values are depicted).